Abstract
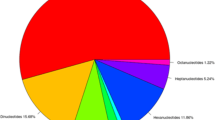
Faba bean (Vicia faba L.) is an important food legume crop with a huge genome. Development of genetic markers for faba bean is important to study diversity and for molecular breeding. In this study, we used Next Generation Sequencing (NGS) technology for the development of genomic simple sequence repeat (SSR) markers. A total of 14,027,500 sequence reads were obtained comprising 4,208 Mb. From these reads, 56,063 contigs were assembled (16,367 Mb) and 2138 SSRs were identified. Mono and dinucleotides were the most abundant, accounting for 57.5 % and 20.9 % of all SSR repeats, respectively. A total of 430 primer pairs were designed from contigs larger than 350 nucleotides and 50 primers pairs were tested for validation of SSR locus amplification. Nearly all (96 %) of the markers were found to produce clear amplicons and to be reproducible. Thirty-nine SSR markers were then applied to 46 faba bean accessions from worldwide origins, resulting in 161 alleles with 87.5 % polymorphism, and an average of 4.1 alleles per marker. Gene diversity (GD) of the markers ranged from 0 to 0.48 with an average of 0.27. Testing of the markers showed that they were useful in determining genetic relationships and population structure in faba bean accessions.


Similar content being viewed by others
References
Abdelkrim J, Robertson BC, Stanton JA, Gemmell NJ (2009) Fast cost-efective development of species-specific microsatalite markers by genomic sequencing. Biotechniques 46(3):185–192. doi:10.2144/000113084
Abid G, Mingeot D, Udupa SM, Muhovski Y, Watillon B, Sassi K, M’hamdi M, Souissi F et al (2015) Genetic relationship and diversity analysis of faba bean (Vicia faba L. var. Minor) genetic resources using morphological and microsatellite molecular markers. Plant Mol Biol Rep 33(6):1755–1767. doi:10.1007/s11105-015-0871-0
Akash MW, Mayers GO (2012) The development of faba bean expressed sequence tag–simple sequence repeats (EST-SSRs) and their validity in diversity analysis. Plant Breed 131(4):522–530. doi:10.1111/j.1439-0523.2012.01969.x
Baloch FS, Karakoy T, Demirbas A, Toklu F, Ozkan H, Hatipoglu R (2014) Variation of some seed mineral contents in open pollinated faba bean (Vicia faba L.) landraces from Turkey. Turk J Agric For 38:591–602. doi:10.3906/tar-1311-31
Cardle L, Ramsay L, Milbourne D, Macaulay M, Marshall D, Waugh R (2000) Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics 156(2):847–854
Cordoba JM, Chavarro C, Schlueter JA, Jackson SA, Blair MW (2010) Integration of physical and genetic maps of common bean through BAC-derived microsatellite markers. BMC Genomics 11:436. doi:10.1186/1471-2164-11-436
Doyle JJ, Doyle JE (1990) Isolation of plant DNA from fresh plant tissue. Focus 12:13–15
Duc G (1997) Faba bean (Vicia faba L.). Field Crop Res 53:99–109
Duc G, Bao S, Baum M, Redden B, Sadiki S, Suso MJ, Vishniakova M, Zong X (2010) Diversity maintenance and use of Vicia faba L. genetic resources. Field Crop Res 115:270–278. doi:10.1016/j.fcr.2008.10.003
Ellwood SR, Phan HTT, Jordan M, Hane J, Torres AM, Avila CM, Cruz- Izquierdo S, Oliver RP (2008) Construction of a comparative genetic map in faba bean (Vicia faba L.); conservation of genome structure with Lens culinaris. BMC Genomics 9:380. doi:10.1186/1471-2164-9-380
Earl DA, VonHolt BM (2012) Structure Harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour 4:359–361. doi:10.1007/s12686-011-9548-7
El-Rodeny W, Kimura M, Hirakawa H, Sabah A, Shirasawa K, Sato S, Tabata S, Sasamoto S et al (2014) Development of EST-SSR markers and construction of a linkage map in faba bean (Vicia faba). Breed Sci 64(3):252–263. doi:10.1270/jsbbs.64.252
FAOSTAT 2014. Crops. Available online: http://faostat3.fao.org
Gong YM, Xu SC, Mao WH, Hu QZ, Zhang GW, Ding J, Li ZY (2010) Generation and characterization of 11 novel est derived microsatellites from Vicia faba (Fabaceae). Am J Bot 97:e69–e71. doi:10.3732/ajb.1000166
Gong YM, Xu SC, Mao WH, Lize Y, Hu QZ, Zhang GW, Ding J (2011) Genetic diversity analysis of faba bean (Vicia faba L.) based on EST-SSR markers. Agric Sci China 10:838–844. doi:10.1016/S1671-2927(11)60069-2
Gresta F, Giovanni A, Emidio A, Lorenzo R, Valerio A (2010) A study of variability in the Sicilian faba bean landrace ‘Larga di Leonforte’. Genet Resour Crop Evol 57(4):523–531. doi:10.1007/s10722-009-9490-7
Hamada H, Petrino MG, Kakunaga T (1982) A novel repeated element with Z-DNA forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A 79(21):6465–6469
Kaur S, Cogan NOI, Forster JW, Paull JG (2014) Assessment of genetic diversity in faba bean based on single nucleotide polymorphism. Diversity 6(1):88–101. doi:10.3390/d6010088
Kaur S, Pembleton LW, Cogan NOI, Savin KW, Leonforte T, Paull J, Materne M, Forster JW (2012) Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers. BMC Genomics 13:104. doi:10.1186/1471-2164-13-104
Koressaar T, Remm M (2007) Enhancement and modifications of primer design program Primer3. Bioinformatics 23(10):1289–1291. doi:10.1093/bioinformatics/btm091
Langmead B, Salzberg SL (2012) Fast gapped – read alignment with Bowtie2. Nat Methods 9(4):357–359. doi:10.1038/nmeth.1923
Leclercq S, Rivals E, Jarne P (2007) Detecting microsatellites within genomes: significant variation among algorithms. BMC Bioinf 8:125. doi:10.1186/1471-105-8-125
Link W, Dixkens C, Singh M, Schwall M, Melchinger AE (1995) Genetic diversity in European and Mediterranean faba bean germ plasm revealed by RAPD markers. Theor Appl Genet 90:27–32. doi:10.1007/BF00220992
Ma Y, Yang T, Guan J, Wang S, Wang H, Sun X, Zong X (2011) Development and characterization of 21 EST-derived microsatellite markers in Vicia faba (fava bean). Am J Bot 98:e22–e24. doi:10.3732/ajb.1000407
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetJ 17(1):10–12, doi:10.14806/ej.17.1.200
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A 70(12):3321–3323. doi:10.1073/pnas.70.12.3321
Ouji A, El-Bok S, Syed NH, Abdellaoui R, Rouaissi M, Flavell AJ, El-Gazzah M (2012) Genetic diversity of faba bean (Vicia faba L.) populations revealed by sequence specific amplified polymorphism (SSAP) markers. Afr J Biotechnol 11:2162–2168. doi:10.5897/AJB11.2991
Pozarkova D, Koblizkova A, Roman B, Torres AM, Lucretti S, Lysak M, Dolezel J, Macas J (2002) Development and characterization of microsatellite markers from chromosome 1-specifi c DNA libraries of Vicia faba. Biol Plant 45:337–345
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Roldan-Ruiz I, Dendauw J, Bockstaele EV, Depicker A, Loose MD (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breed 6:125–134. doi:10.1023/A:1009680614564
Senan S, Kizhakayil D, Sasikumar B, Sheeja TE (2014) Methods for development of microsatellite markers: an overview. Not Sci Biol 6(1):1–13. doi:10.15835/nsb.6.1.9199
Shendure J, Ji H (2008) Next-generation DNA sequencing. Nat Biotechnol 26:1135–1145. doi:10.1038/nbt1486
Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19(6):1117–1123. doi:10.1101/gr.089532.108
Suresh S, Park JH, Cho GT, Lee HS, Baek HG, Lee SY, Chung JW (2013) Development and molecular characterization of 55 novel polymorphic cDNA-SSR markers in faba bean (Vicia faba L.) using 454 pyrosequencing. Molecules 18:1844–1856. doi:10.3390/molecules18021844
Terzopoulos PJ, Bebeli PJ (2008) Genetic diversity analysis of Mediterranean faba bean (Vicia faba L.) with ISSR markers. Field Crop Res 108:39–44. doi:10.1016/j.fcr.2008.02.015
Torres AM, Roman B, Avila CM, Satovic Z, Rubiales D, Sillero JC, Cubero JI, Moreno MT (2006) Faba bean breeding for resistance against biotic stresses: towards application of marker technology. Euphytica 147(1):67–80. doi:10.1007/s10681-006-4057-6
Toth G, Gaspari Z, Jurka J (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res 10:967–981. doi:10.1101/gr.10.7.967
Van de Ven M, Powell W, Ramsay G, Waugh R (1990) Restriction fragment length polymorphisms as genetic markers in Vicia. Heredity 65:329–342. doi:10.1038/hdy.1990.102
Wang HF, Zong XX, Guan JP, Yang T, Sun XL, Ma Y, Redden R (2012) Genetic diversity and relationship of global faba bean (Vicia faba L.) germplasm revealed by ISSR markers. Theor Appl Genet 124(5):789–797. doi:10.1007/s00122-011-1750-1
Yang T, Bao SY, Ford R, Jia TJ, Guan JP, He YH, Sun XL, Jiang JY et al (2012) High-throughput novel microsatellite marker of faba bean via next generation sequencing. BMC Genomics 13:602. doi:10.1186/1471-2164-13-602
Zalapa JE, Cuevas H, Zhu H, Steffan S, Senalik D, Zeldin E, McCown B, Harbut R, Simon P (2012) Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Am J Bot 99(2):193–208. doi:10.3732/ajb.1100394
Zane L, Bargelloni L, Patarnello T (2002) Strategies for microsatellite isolation: a review. Mol Ecol 11(1):1–16. doi:10.1046/j.0962-1083.2001.01418.x
Zeid M, Schon CC, Link W (2003) Genetic diversity in recent elite faba bean lines using AFLP markers. Theor Appl Genet 107(7):1304–1314. doi:10.1007/s00122-003-1350-9
Zeid M, Mitchell S, Link W, Carter M, Nawar A, Fulton T, Kresovich S (2009) Simple sequence repeats (SSRs) in faba bean: new loci from Orobanche-resistant cultivar Giza 402. Plant Breed 128(2):149–155. doi:10.1111/j.1439-0523.2008.01584.x
Zong X, Liu X, Guan J, Wang S, Liu Q, Paull JG, Redden R (2009) Molecular variation among Chinese and global winter faba bean germplasm. Theor Appl Genet 118(5):971–978. doi:10.1007/s00122-008-0954-5
Acknowledgments
This study was supported by grant 0424.STZ.2013-2 from The Republic of Turkey’s Ministry of Science, Industry and Technology to AF with contributions from Polen Seed Co. Additional support was received from the Scientific and Technological Research Council of Turkey grant 113E326 to JA. We are grateful to Ali Uncu and Ayse Ozger Uncu for SSR validation by sequencing.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
Supplementary Table S1 Preprocessing and assembly statistics for the faba bean genomic sequences. Supplementary Table S2 Primer details for the 11 SSR markers not given in Table 3. Supplementary Table S3 Faba bean genomic sequence displaying the four simple sequence repeat motifs within the PCR amplicons. Supplementary Fig. S1 Bar plot showing genetic structure of 46 faba bean accessions with K = 2. Each accession is represented by a vertical bar which is partitioned according to estimated membership in the two clusters. Cluster A fraction is shown in red and cluster B fraction in green. (DOCX 123 kb)
Rights and permissions
About this article
Cite this article
Abuzayed, M.A., Goktay, M., Allmer, J. et al. Development of genomic simple sequence repeat markers in faba bean by next-generation sequencing. Plant Mol Biol Rep 35, 61–71 (2017). https://doi.org/10.1007/s11105-016-1003-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-016-1003-1