Abstract
Background
Faba bean (Vicia faba L.) is an important food legume crop, grown for human consumption globally including in China, Turkey, Egypt and Ethiopia. Although genetic gain has been made through conventional selection and breeding efforts, this could be substantially improved through the application of molecular methods. For this, a set of reliable molecular markers representative of the entire genome is required.
Results
A library with 125,559 putative SSR sequences was constructed and characterized for repeat type and length from a mixed genome of 247 spring and winter sown faba bean genotypes using 454 sequencing. A suit of 28,503 primer pair sequences were designed and 150 were randomly selected for validation. Of these, 94 produced reproducible amplicons that were polymorphic among 32 faba bean genotypes selected from diverse geographical locations. The number of alleles per locus ranged from 2 to 8, the expected heterozygocities ranged from 0.0000 to 1.0000, and the observed heterozygosities ranged from 0.0908 to 0.8410. The validation by UPGMA cluster analysis of 32 genotypes based on Nei's genetic distance, showed high quality and effectiveness of those novel SSR markers developed via next generation sequencing technology.
Conclusions
Large scale SSR marker development was successfully achieved using next generation sequencing of the V. faba genome. These novel markers are valuable for constructing genetic linkage maps, future QTL mapping, and marker-assisted trait selection in faba bean breeding efforts.
Similar content being viewed by others
Background
Faba bean (Vicia faba L.) is an important temperate legume, grown for human consumption and animal feed due to its high protein and fibre content [1, 2]. The crop also replaces available nitrogen in the soil when used in rotation with cereals and oilseeds, and thus is expected to be a highly beneficial component in future temperate Low Carbon Agricultural systems. China is the largest faba bean producer (40.36%) with an average dry grain production (2005–2009) of 1,720,000 metric tonnes (mt) from 945,400 hectares; followed by Ethiopia (476,026 mt), France (331,122 mt), Egypt (274,040 mt) and Australia (196,800 mt) [3].
However, faba bean suffers from several major biotic and abiotic factors that constrain productivity. Although significant genetic gain to overcome these has been made through traditional breeding practices [1], progress through the use of genomics and associated biotechnologies is limited. This is due mainly to the large genome size (13GB; [4]), which is approximately 25 times larger than that of the model legume Medicago truncatula, and 2.5 times larger than Pisum sativum[1], together with a lack of financial investment in this crop species.
Recent advances in next generation sequencing (NGS) technologies enable the generation of large volumes of sequence efficiently and cost-effectively [5, 6]. This has led to a revolution in biological and agricultural applications including identification of genes correlated with key breeding traits through high-density SNP marker and genome-wide association analysis studies (GWAS) [7, 8]. Another outcome is the ability to accurately identify sequences flanking simple sequence repeat (SSR) regions for use as locus-specific markers for downstream genotyping. Otherwise known as microsatellites, SSRs are tandemly repeated motifs of 1 to 6 nucleotides found in both coding and non-coding regions [9, 10]. These have become a marker of choice in many genotyping applications due to their relatively high abundance, high level of allelic variation, co-dominant inheritance, analytical simplicity and transferability of results across laboratories [11].
A limited number of characterized SSR loci (<120) which have been validated over relatively few genetic backgrounds are available for faba bean. Initially, Pozarkova et al. developed primers to 25 SSR loci detected in chromosome 1 DNA libraries [12]. Subsequently, Zeid et al. developed primers to 54 SSR loci [13] and Gong et al. developed 11 EST-SSR loci primers [14]. Most recently, EST sequences within the public domain databases were screened and an additional 21 novel SSR loci were characterized and validated among 32 faba bean accessions [15].
Besides providing a cost-effective valuable source for molecular marker generation, the identification of SSR within ESTs is an effective approach for gene discovery and transcript pattern characterization, particularly if through mapping an EST-SSR or EST marker is significantly associated with a QTL [16–18]. This may be achieved by searching for SSR associated sequences within EST of a well characterised crop or model plant species. Together with the advantage of in silico analysis, this approach has the potential to substantially broaden the field of comparative studies to species where limited or no sequence information is available.
The present study identified high-quality putative SSR loci and flanking primer sequences cheaply and efficiently using the Roche 454 GS FLX Titanium platform. The resultant SSR sequences were characterized and validated through successful amplification of randomly selected target loci across a selection of faba bean genotypes from diverse geographic origin.
Methods
Plant material
A total of 247 faba bean accessions were selected from the National Genebank of China held at the Institute of Crop Science (ICS), Chinese Academy of Agricultural Sciences (CAAS), Beijing. Of these, 100 originated from China, 54 were from other Asian countries, 39 were from Europe, 30 were from Africa, 14 were from the America, 9 breeding lines were sourced from the ICARDA (International Center for Agricultural Research in the Dry Areas) faba bean breeding program and one was from Oceania (Additional file 1: Table S1).
DNA isolation, library preparation and 454 sequencing
Seven days after seed were left on moist filter paper in the dark at 22°C, sprouts from each of the 247 genotypes were collected. A single sprout of each genotype and of approximately the same weight was pooled and total gDNA was extracted using the CTAB method [19, 20].
Genome libraries were constructed using eight biotin labeled probes and a selective hybridization with streptavidin coated bead method [21–23]. The probes were: pGA, pAC, pAAT, pAAC, pAAG, pATGT, pGATA and pAAAT. The quality of libraries was inspected by randomly selecting and sequencing 276 clones. The cloning vector was pEASY-T1 (TransGen Biotechnology Co., Ltd), and the primers used for sequencing were F: 5′-GTAAAACGACGGCCAGT-3′ and R: 5′-CAGGAAACAGCTATGAC-3′. Libraries were considered to be of high quality if the length of sequences were from 200 to 1000 bp, as evidenced on agarose gel.
Subsequently, entire libraries were equally pooled and subjected to 454 sequencing with GS-FLX Titanium reagents at Beijing Autolab Biotechnology Co., Ltd (China). All processing and analyses of the sequencing data was performed with GS-FLX Software v2.0.01 (454 Life Sciences, Roche, Germany). Using a series of normalization, correction and quality-filtering algorithms, the 454 sequencing data were processed to screen and filter for weak signals and low-quality reads, and to trim the read ends for 454 adaptor sequences using the EMBOSS [24] software package. The sequencing data were then submitted to the ′National Center for Biotechnology Information (NCBI) short read archive and given the accession number SRP006387.
SSR loci search and primer design
The software MISA (Mi crosa tellite identification) tool (http://pgrc.ipk-gatersleben.de/misa/) was configured to locate a minimum of 10 bp: monomers (×10), 2-mers (×6), 3-mers (×5), 4-mers (×5), 5-mers (×5) and 6-mers (×5). This tool allowed the identification and localization of perfect microsatellites as well as compound microsatellites. The maximum size of interruption allowed between two different SSR in a compound sequence was 100 bp. Subsequently, Primer 3.0 (http://www-genome.wi.mit.edu/genome_software/other/primer3.html.) was used to design primer pairs to the flanking sequences of each unique SSR.
SSR characterization and validation
The number of different types of SSR, length (motif bp × number of motifs) and SSR position was searched and analyzed for using a bespoke program written in MISA files [25] and plotted by OpenOffice.org Calc.
Marker assessment
Polymerase chain reactions (PCR) were performed in 20 μl reaction volumes containing 0.5 U of Taq DNA polymerase (Zhexing, Beijing, China), 1 × PCR BufferII, 1.5 mM MgCl2, 25 μM of dNTP, 0.4 μM primer, and 50 ng of genomic DNA. Microsatellite loci were amplified on a Heijingang Thermal Cycler (Eastwin, Beijing, China) with the following cycle: 5 min initial denaturation at 95°C; 35 cycles of 30s at 95°C, 30s at the optimized annealing temperature (Table 1), 45s of elongation at 72°C, and a final extension at 72°C for 10min. PCR products were initially assessed for size polymorphism on 6% denaturing polyacrylamide gels and visualized by silver nitrate staining.
The genotyping data was subsequently used to determine genetic relationships among 32 V. faba accessions (eleven from China, seven from Asia, five from Europe, five from Africa, three from the Americas and one from Oceania; (Additional file 1: Table S1). The number of alleles (Na), expected (He) heterozygosities and observed (Ho) heterozygosities were calculated using POPGEN1.32 [26]. The cluster analysis of 32 genotypes was carried out based on Nei's unbiased measures of genetic distance [27] by using the unweighted pair-group method with arithmetic average (UPGMA), and the dendrogram was drawn by MEGA4 [28].
Results
Quality inspection of the DNA library
The recombination rate within the constructed SSR-enriched V. faba library was 73.9%. Among the 276 clones sequenced, 31.9% contained SSR sequences within an insert that ranged from 0.2 to 1.0 kb in size.
454 sequencing and characterization reads
A total of 578,251 reads were generated from the pooled library, and 532,599 read sequences were used for further analysis after adaptor removal. Adenine was the most abundant nucleotide (30%), followed by thymine (27%), guanine (22%) and cytosine (21%). The mean GC content was 43%. The average length of read sequence was 305 bp, with a maximum length of 635 bp (Figure 1).
Identification of SSR loci
After MISA analysis, the number of sequences containing an SSR was 125,559, and in total 250,393 SSR loci were detected. The number of sequences containing more than one SSR loci was 61,266 and the number of SSRs present in compound formation was 122,988 (Table 1).
The total size of SSR motif sequences was 8,759,185 bp, with an average motif length of 69 bp. Of these, 25% comprised more than one discrete repeat and a high proportion (49%) was located within compound repeats. The majority of identified SSR motifs (83%) were located between the 5’-terminus and mid regions of the cloned sequences, and within 200 bp of the 5’-terminus (Figure 2). A total of 28,503 primer pairs were designed for future assessment of locus amplification (Additional file 2: Table S2).
Abundance and length frequencies of SSR repeat motifs
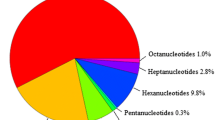
The most common SSR motifs comprised trinucleotide and dinucleotide repeats (Figure 3). The majority of the trinucleotide repeats were from 15 to 30 bp in length. Within the 1,188 characterised mononucleotide SSR, (A/T)n was almost three times more common than (C/G)n, particularly at the 11–12 bp length. The dinucleotide repeats (AC/GT)n and (AG/CT)n were predominant, representing 99.2% of all of the dinucleotides characterised. Triucleotide (AAC/GTT)n repeats were the most abundant (96.5%). Twenty two unique tetranucleotide repeat motifs were identified, with the most common being AGAT/ATCT (66.4%), ACAG/CTGT (19.3%) and ACAT/ATGT (9.1%). Pentanucleotide and hexanucleotide motifs were far less frequent, together comprising only 0.1% of the total SSR detected. The dominant pentanucelotide motif was AGAGT/ATCTC (23.8%) and the most common hexanucelotide motif was ACACGC/CGTGTG (49.5%) (Additional files 3, 4, 5, 6, 7 and 8: Figure S1-S6).
Compound SSR analysis
Two types of compound SSR were identified; those without an interruption between two motifs (ie (CA)12(ACG)37 and noted as C* type) and those with an interruption between two motifs ( ie (AAC)7gtcaat(AAC)5 and noted as C type). In total, 1,893 C* type and 59,369 C type compound SSR loci were detected among those sequenced, reflecting the complexity of the faba bean genome.
Validation of SSR assay
Of the 150 primer pairs selected for validation of SSR locus amplification, 102 produced a reproducible and clear amplicon of the expected size. Of these, 94 (63%) were polymorphic among thirty-two genotypes assessed (Table 2). The number of alleles per locus ranged from 2 to 8, the expected heterozygosities ranged from 0.0000 to 1.0000, and the observed heterozygosities ranged from 0.0908 to 0.8410 (Table 3).
The dendrogram showed that the 32 faba bean genotypes fell into four distinct clusters (Figure 4). Cluster 1 comprised accessions from China and other Asian countries except for one accessions from Africa. Cluster 2 comprised accessions from Europe and nearby regions such as Syria. Cluster 3 comprised accessions from Africa and Cluster 4 contained accessions from America, Oceania and Africa. The pattern of diversity was similar to that previously observed using AFLP [29] and ISSR [30] markers.
Discussion
This study demonstrated that massively parallel sequencing technology offers opportunity to quickly identify large numbers of high quality SSR with diverse motifs from a genetically orphaned species such as Vicia faba. Given the huge number of marker loci identified in this study, future SSR marker optimisation may be best focussed on those comprising trinucleotide repeats. These repeats are generally more robust since they are reported to give fewer “stutter bands” than those based on dinucleotide repeats [31, 32]. Also, trinucleotide repeats in particular have been demonstrated to be highly polymorphic and stably inherited in the human genome [33–35]. While the tri- and dinucleotide repeats mostly contributed to the major proportion of SSRs, a very small share was contributed by mono-, tetra-, penta- and hexa-nucleotide repeats. A similar trend was observed in other species [36].
The conversion of SSR-containing sequences into single locus markers may have a low success rate due to complex and/or insufficient flanking sequence. For example, just 20% of the identified dinucleotide repeats from spruce were converted to clear, discrete markers [37]. Similar observations were made for pine [38], wheat [39] and previously for V. faba[12]. Another factor affecting the development of clear markers is the complexity of the repeat motifs, indeed a high proportion of the SSR in the current study comprised compound repeats (49.1%). Nevertheless, this study has provided the selected data required to potentially develop tens of thousands of novel SSR markers for the faba bean genome.
Previously, a total of 304,680 reads were generated and 802 EST-SSR primer pairs were designed from transcriptome sequencing of faba bean [40]. From this, 81 primer pairs were developed, of which 48% produced polymorphic markers on the genotypes assessed. In our study, 68% (102) of the SSR loci identified were accurately amplified, of which 63% (94) were polymorphic among the genotypes tested. This may be indicatative of the larger number of SSR loci detected, inclusive of non-transcribed sequences. Hence these markers may be more representative of the entire genome for the purposes of germplasm diversity assessment and conservation purposes [41]. Meanwhile, the identification of EST-SSR within sequences provides future opportunity to mine the expressed sequences for significant physical and functional association with traits of interest in marker-assisted faba bean breeding.
Conclusion
This work represents a major advance in the identification of large numbers of informative SSR loci in V. faba by application of 454 GS FLX Titanium sequencing technology.
Abbreviations
- SSR:
-
Simple sequence repeat
- QTL:
-
Quantative Trait Locus
- MAS:
-
Marker-assisted selection
- NGS:
-
Next generation sequencing
- EST:
-
Express sequence tag
- NCBI:
-
National Center for Biotechnology Information
- CTAB:
-
Cetyltrimethylammonium bromid
- MISA:
-
Microsatellite identification
- Na :
-
Number of alleles
- He :
-
Expected heterozygosities
- Ho :
-
Observed heterozygosities.
References
Rispail N, Kal P, Kiss GB, Ellis THN, Gallardo K, Thompson RD, Prats E, Larrainzar E, Ladrera R, Gonzalez EM, ArreseIgor C, Ferguson BJ, Gresshoff PM, Rubiales D: Model legumes contribute to faba bean breeding. Field Crop Res. 2010, 115: 253-269. 10.1016/j.fcr.2009.03.014.
Duc G, Marget P, Esnault R, Le Guen J, Bastianelli D: Genetic variability for feeding value of faba bean seeds (Vicia faba): Comparative chemical composition of isogenics involving zero-tannin and zero-vicine genes. J Agric Sci. 1999, 133: 185-196. 10.1017/S0021859699006905.
FAOSTAT: 2010,http://faostat.fao.org/site/291/default.aspx,
Johnston JS, Bennett MD, Rayburn AL, Galbraith DW, Price HJ: Reference standards for determination of DNA content of plant nuclei. Am J Bot. 1999, 86: 609-10.2307/2656569.
Morozova O, Marra MA: Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008, 92: 255-264. 10.1016/j.ygeno.2008.07.001.
Mardis ER: The impact of next-generation sequencing technology on genetics. Trends Genet. 2008, 24: 133-141. 10.1016/j.tig.2007.12.007.
Lam HM, Xu X, Liu X, Chen W, Yang G, Wong FL, Li MW, He W, Qin N, Wang B, Li J, Jian M, Wang J, Shao G, Wang J, Sun SSM, Zhang G: Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 2010, 42: 1053-1059. 10.1038/ng.715.
Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, Li M, Fan D, Guo Y, Wang A, Wang L, Deng L, Li W, Lu Y, Weng Q, Liu K, Huang T, Zhou T, Jing Y, Li W, Lin Z, Buckler ES, Qian Q, Zhang QF, Li J, Han B: Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010, 42: 961-967. 10.1038/ng.695.
Tautz D, Renz M: Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984, 12: 4127-4138. 10.1093/nar/12.10.4127.
Lagercrantz U, Ellegren H, Andersson L: The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Res. 1993, 21: 1111-1115. 10.1093/nar/21.5.1111.
Rafalski JA, Tingey SV: Genetic diagnostics in plant breeding: RAPDs, microsatellites and machines. Trends Genet. 1993, 9: 275-280. 10.1016/0168-9525(93)90013-8.
Požárková D, Koblížková A, Román B, Torres AM, Lucretti S, Lysák M, Doležel J, Macas J: Development and characterization of microsatellite markers from chromosome 1-specific DNA libraries of Vicia faba. Biologia Plantarum. 2002, 45: 337-345. 10.1023/A:1016253214182.
Zeid M, Mitchell S, Link W, Carter M, Nawar A, Fulton T, Kresovich S: Simple sequence repeats (SSRs) in faba bean: new loci from Orobanche-resistant cultivar ‘Giza 402’. Plant Breeding. 2009, 128: 149-155. 10.1111/j.1439-0523.2008.01584.x.
Gong Y-M, Xu S-C, Mao W-H, Hu Q-Z, Zhang G-W, Ding J, Li Z-Y: Generation and characterization of 11 novel EST derived microsatellites from Vicia faba (Fabaceae). Am J Bot. 2010, 97: e69-e71. 10.3732/ajb.1000166.
Ma Y, Yang T, Guan J, Wang S, Wang H, Sun X, Zong X: Development and characterization of 21 EST-derived microsatellite markers in Vicia faba (fava bean). Am J Bot. 2011, 98: e22-e24. 10.3732/ajb.1000407.
Zhang WK, Wang YJ, Luo GZ, Zhang JS, He CY, Wu XL, Gai JY, Chen SY: QTL mapping of ten agronomic traits on the soybean (Glycine max L. Merr.) genetic map and their association with EST markers. Theor Appl Genet. 2004, 108: 1131-1139. 10.1007/s00122-003-1527-2.
Matthews BF, Devine TE, Weisemann JM, Beard HS, Lewers KS, MacDonald MH, Park Y-B, Maiti R, Lin J-J, Kuo J, Pedroni MJ, Cregan PB, Saunders JA: Incorporation of sequenced cDNA and genomic markers into the soybean genetic map. Crop Science. 2001, 41: 516-521. 10.2135/cropsci2001.412516x.
Hisano H, Sato S, Isobe S, Sasamoto S, Wada T, Matsuno A, Fujishiro T, Yamada M, Nakayama S, Nakamura Y, Watanabe S, Harada K, Tabata S: Characterization of the soybean genome using EST-derived microsatellite markers. DNA Res. 2007, 14: 271-281.
Dellaporta S, Wood J, Hicks J: A plant DNA minipreparation: Version II. Plant Molecular Biology Reporter. 1983, 1: 19-21. 10.1007/BF02712670.
Doyle JJ, Doyle JL: A rapid total DNA preparation procedure for fresh plant tissue. Focus. 1990, 12: 13-15.
Kandpal RP, Kandpal G, Weissman SM: Construction of libraries enriched for sequence repeats and jumping clones, and hybridization selection for region-specific markers. Proc Natl Acad Sci USA. 1994, 91: 88-92. 10.1073/pnas.91.1.88.
Armour JA, Neumann R, Gobert S, Jeffreys AJ: Isolation of human simple repeat loci by hybridization selection. Hum Mol Genet. 1994, 3: 599-565. 10.1093/hmg/3.4.599.
Glenn TC, Schable NA: Isolating microsatellite DNA loci. Methods Enzymol. 2005, 395: 202-222.
Rice P, Longden I, Bleasby A: EMBOSS: the European molecular biology open software suite. Trends Genet. 2000, 16: 276-277. 10.1016/S0168-9525(00)02024-2.
Thiel , Michalek , Varshney , Graner : Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet. 2003, 106: 411-422.
Yeh FC, Boyle TJB: Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belgian Journal of Botany. 1997, 129: 157-
Nei M: Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978, 89: 583-590.
Tamura K, Dudley J, Nei M, Kumar S: MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007, 24: 1596-1599. 10.1093/molbev/msm092.
Zong X, Liu X, Guan J, Wang S, Liu Q, Paull JG, Redden R: Molecular variation among Chinese and global winter faba bean germplasm. Theor Appl Genet. 2009, 118: 971-978. 10.1007/s00122-008-0954-5.
Wang HF, Zong XX, Guan JJ, Yang T, Sun XL, Ma Y, Redden R: Genetic diversity and relationship of global faba bean (Vicia faba L.) germplasm revealed by ISSR markers. Theor Appl Genet. 2012, 124: 789-797. 10.1007/s00122-011-1750-1.
Hearne CM, Ghosh S, Todd JA: Microsatellites for linkage analysis of genetic traits. Trends Genet. 1992, 8: 288-294.
Diwan N, Cregan PB: Automated sizing of fluorescent-labeled simple sequence repeat (SSR) markers to assay genetic variation in soybean. Theor Appl Genet. 1997, 95: 723-733. 10.1007/s001220050618.
Edwards A, Civitello A, Hammond HA, Caskey CT: DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am J Hum Genet. 1991, 49: 746-756.
Gastier JM, Pulido JC, Sunden S, Brody T, Buetow KH, Murray JC, Weber JL, Hudson TJ, Sheffield VC, Duyk GM: Survey of trinucleotide repeats in the human genome: assessment of their utility as genetic markers. Hum Mol Genet. 1995, 4: 1829-1836. 10.1093/hmg/4.10.1829.
Sheffield VC, Weber JL, Buetow KH, Murray JC, Even DA, Wiles K, Gastier JM, Pulido JC, Yandava C, Sunden SL, Mattes G, Businga T, McClain A, Beck J, Scherpler T, Gilliam J, Zhong J, Duyk GM: A collection of tri- and tetranucleotide repeat markers used to generate high quality, high resolution human genome-wide linkage maps. Hum Mol Genet. 1995, 4: 1837-1844. 10.1093/hmg/4.10.1837.
Sonah H, Deshmukh RK, Sharma A, Singh VP, Gupta DK, Gacche RN, Rana JC, Singh NK, Sharma TR: Genome-wide distribution and organization of microsatellites in plants: an insight into marker development in Brachypodium. PLoS One. 2011, 6: e21298-10.1371/journal.pone.0021298.
Pfeiffer A, Olivieri AM, Morgante M: Identification and characterization of microsatellites in Norway spruce (Picea abies K.). Genome. 1997, 40: 411-419. 10.1139/g97-055.
Smith D, Devey ME: Occurrence and inheritance of microsatellites in Pinus radiata. Genome. 1994, 37: 977-983. 10.1139/g94-138.
Roder MS, Plaschke J, Konig SU, Borner A, Sorrells ME, Tanksley SD, Ganal MW: Abundance, variability and chromosomal location of microsatellites in wheat. Mol Gen Genet. 1995, 246: 327-333. 10.1007/BF00288605.
Kaur S, Pembleton LW, Cogan NO, Savin KW, Leonforte T, Paull J, Materne M, Forster JW: Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers. BMC Genomics. 2012, 13: 104-10.1186/1471-2164-13-104.
Zong X, Redden RJ, Liu Q, Wang S, Guan J, Liu J, Xu Y, Liu X, Gu J, Yan L, Ades P, Ford R: Analysis of a diverse global Pisum sp. collection and comparison to a Chinese local P. sativum collection with microsatellite markers. Theor Appl Genet. 2009, 118: 193-204. 10.1007/s00122-008-0887-z.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 31101198), the China Agriculture Research System (CARS-09) from the Ministry of Agriculture of China and Social Development of Science and Technology Plan (no. 2010BB007) from Yunnan Government. We are grateful to Dr. Dahai Wang and Liping Sun (Beijing Autolab Biotechnology Co., Ltd) for their special contribution to this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TY performed bioinformatic analysis, primer design and drafted the manuscript. SYB created the SSR sequences rich DNA library, and participated in 454 sequencing. RF assisted in designing experiment and preparing the manuscript. TJJ tested SSR markers. JPG and YHH prepared all the seeds of V. faba. XLS and JYJ took charge of quality inspection of the DNA library. JJH and XYZ participated in conceiving the study and the manuscript drafting. XXZ designed and coordinated the study, and assisted in preparing the manuscript. All authors read and approved the final manuscript.
Tao Yang, Shi-ying Bao contributed equally to this work.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Yang, T., Bao, Sy., Ford, R. et al. High-throughput novel microsatellite marker of faba bean via next generation sequencing. BMC Genomics 13, 602 (2012). https://doi.org/10.1186/1471-2164-13-602
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-13-602