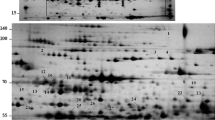
Abstract
Plants respond to UV stress by producing antioxidant molecules and by altering their metabolism through the regulation of specific gene family members. Globe artichoke (Cynara cardunculus var. scolymus L.—Compositae family) is an attractive model species for studying the protein networks involved in UV response, being characterised by high loads of antioxidants, which are boosted by UV-C rays. The effect of UV-C on leaf proteome was investigated 24 h after irradiation by applying a polyethylene glycol (PEG) fractionation method, which we previously developed to cope with the so-called high abundance proteins, followed by two-dimensional electrophoresis analysis. Thirty-eight UV-C modulated proteins were analysed by means of a nano-liquid chromatography–nanospray tandem mass spectrometry, and 27 were successfully identified using a customised Compositae protein database, including a set of 19,055 globe artichoke unigenes. Proteins were categorised according to Gene Ontology (GO) and visualised on their cellular pathway: Most of them were involved not only in primary metabolism (e.g. photosynthesis) and in abiotic stress responses (e.g. reactive oxygen species scavenging) but also in cell wall modification, secondary metabolism, and regulation of transcription. To date, this is the first report on globe artichoke proteomic expression analysis following abiotic stress. Our results confirm the effectiveness of the PEG fractionation method in excluding most of the ribulose bisphosphate carboxylase/oxygenase and highlighting some major occurring phenomena.




Similar content being viewed by others
References
Acquadro A, Falvo S, Mila S, Albo A, Comino C, Moglia A, Lanteri S (2009) Proteomics in globe artichoke: protein extraction and sample complexity reduction by PEG fractionation. Electrophoresis 30(9):1594–1602
Ahsan N, Lee D, Alam I et al (2008) Comparative proteomic study of arsenic-induced differentially expressed proteins in rice roots reveals glutathione plays a central role during As stress. Proteomics 8:3561–3576
Albo A, Mila S, Digilio G et al (2007) Proteomic analysis of a genetically modified maize flour carrying Cry1Ab gene and comparison to the corresponding wild-type. Maydica 52:443–455
Bian F, Zheng C, Qu F, Gong X, You C (2010) Proteomic analysis of somatic embryogenesis in Cyclamen persicum Mill. Plant Mol Biol Report 28(1):22–31
Bollenbach T, Sharwood R, Gutierrez R et al (2009) The RNA-binding proteins CSP41a and CSP41b may regulate transcription and translation of chloroplast-encoded RNAs in Arabidopsis. Plant Mol Biol 69:541–552
Braam J (1992) Regulation of expression of calmodulin and calmodulin-related genes by environmental stimuli in plants. Cell Calcium 13:457–463
Braam J, Davis R (1990) Rain-induced, wind-induced, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell 60:357–364
Bradford M (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye-binding. Anal Biochem 72:248–254
Casati P, Zhang X, Burlingame AL, Walbot V (2005) Analysis of leaf proteome after UV-B irradiation in maize lines differing in sensitivity. Mol Cell Proteomics 4:1673–1685
Cho S, Kim J, Park J et al (2006) Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett 580:3136–3144
Conesa A, Gotz S, Garcia-Gomez J, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676
Coohill T (1999) Conventional UV-visible sources. Photochem Photobiol 69:15S
Damerval C, De Vienne D, Zivy MHT (1986) Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling proteins. Electrophoresis 7:52–54
Ding C, You J, Liu Z, Rehmani MIA, Wang S, Li G, Wang Q, Ding Y (2011) Proteomic analysis of low nitrogen stress-responsive proteins in roots of rice. Plant Mol Biol Report. doi:10.1007/s11105-010-0268-z
Dong M, Zhang X, Zhuang Z, Zou J, Ye N, Xu D, Mou S, Liang C, Wang W (2011) Characterization of the LhcSR gene under light and temperature stress in the green alga Ulva linza. Plant Mol Biol Report. doi:10.1007/s11105-011-0311-8
Du HM, Liang Y, Pei KQ, Ma K (2011) UV radiation-responsive proteins in rice leaves: a proteomic analysis. Plant Cell Physiol 52(2):306–316
Feng L, Wang K, Li Y et al (2007) Overexpression of SBPase enhances photosynthesis against high temperature stress in transgenic rice plants. Plant Cell Rep 26:1635–1646
Iliev E, Xu W, Polisensky D et al (2002) Transcriptional and posttranscriptional regulation of Arabidopsis TCH4 expression by diverse stimuli. Roles of cis regions and brassinosteroids. Plant Physiol 130:770–783
Jellouli N, Jouira HB, Daldoul S, Chenennaoui S, Ghorbel A, Salem AB, Gargouri A (2010) Proteomic and transcriptomic analysis of grapevine pr10 expression during salt stress and functional characterization in yeast. Plant Mol Biol Report 28(1):1–8
Kaspar S, Matros A, Mock HP (2010) Proteome and flavonoid analysis reveals distinct responses of epidermal tissue and whole leaves upon UV−B Radiation of barley (Hordeum vulgare L.) seedlings. J Proteome Res 9(5):2402–2411
Kasprzewska A (2003) Plant chitinases—regulation and function. Cell Mol Biol Lett 8:809–824
Kun W, Feng G, Renshan Z, Shaoqing L, Yingguo Z (2011) Expression, purification, and secondary structure prediction of pentatricopeptide repeat protein RF1A from rice). Plant Mol Biol Report. doi:10.1007/s11105-010-0260-7
Kuun K, Okole B, Bornman L (2001) Protection of phenylpropanoid metabolism by prior heat treatment in Lycopersicon esculentum exposed to Ralstonia solanacearum. Plant Physiol Biochem 39:871–880
Lefebvre S, Lawson T, Zakhleniuk O et al (2005) Increased sedoheptulose-1,7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiol 138:451–460
Li D, Staehelin C, Wang W, Peng S (2010) Molecular cloning and characterization of a chitinase-homologous gene from Mikania micrantha infected by Cuscuta campestris. Plant Mol Biol Report 28(1):90–101
Michiels A, Van Laere A, Van den Ende W et al (2004) Expression analysis of a chicory fructan 1-exohydrolase gene reveals complex regulation by cold. J Exp Bot 55:1325–1333
Miyagawa Y, Tamoi M, Shigeoka S (2001) Overexpression of a cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase in tobacco enhances photosynthesis and growth. Nat Biotechnol 19:965–969
Moglia A, Lanteri S, Comino C, Acquadro A, de Vos R, Beekwilder J (2008) Stress-induced biosynthesis of dicaffeoylquinic acids in globe artichoke. J Agric Food Chem 56(18):8641–8649
Moon Y, Wang X, Morris M (2006) Dietary flavonoids: effects on xenobiotic and carcinogen metabolism. Toxicol in Vitro 20:187–210
Petit A, Baillieul F, Vaillant-Gaveau N et al (2009) Low responsiveness of grapevine flowers and berries at fruit set to UV-C irradiation. J Exp Bot 60:1155–1162
Polisensky D, Braam J (1996) Cold-shock regulation of the arabidopsis TCH genes and the effects of modulating intracellular calcium levels. Plant Physiol 11:1271–1279
Qian L, Wenyu Y, Xiaoping Y, Songlin R, Haifeng C, Huasheng M, Zihong Y (2011) A preliminary study on proteome variations associated with gall formation in Zizania latifolia Trucs). Plant Mol Biol Report 29(2):360–368
Ramnani PGE, Bingham M, van Bruggen P, Tuohy KM, Gibson GR (2010) Prebiotic effect of fruit and vegetable shots containing Jerusalem artichoke inulin: a human intervention study. Br J Nutr 1:1–8
Regalado A, Pinheiro C, Vidal S et al (2000) The Lupinus albus class-III chitinase gene, IF3, is constitutively expressed in vegetative organs and developing seeds. Planta 210:543–550
Righetti P, Boschetti E (2007) Sherlock Holmes and the proteome—a detective story. FEBS J 274(4):897–905
Ronkart S, Blecker C, Fourmanoir H et al (2007) Isolation and identification of inulooligosaccharides resulting from inulin hydrolysis. Anal Chim Acta 604:81–87
Scaglione D, Acquadro A, Portis E, Taylor CA, Lanteri S, Knapp SJ (2009) Ontology and diversity of transcript-associated microsatellites mined from a globe artichoke EST database. BMC Genomics 10(1):454
Shevchenko A, Tomas H, Havlis J et al (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860
Singh A, Sarkar A, Singh S et al (2010) Investigation of supplemental ultraviolet-B-induced changes in antioxidative defense system and leaf proteome in radish (Raphanus sativus L. cv Truthful): an insight to plant response under high oxidative stress. Protoplasma 245:75–83
Stapletons A (1992) Ultraviolet-radiation and plants—burning questions. Plant Cell 4:1353–1358
Timperio A, Egidi M, Zolla L (2008) Proteomics applied on plant abiotic stresses: role of heat shock proteins (HSP). J Proteomics 71:391–411
Van den Ende W, De Coninck B, Van Laere A (2004) Plant fructan exohydrolases: a role in signaling and defense? Trends Plant Sci 9:523–528
Wang M, Simon J, Aviles I, He K, Zheng Q, Tadmor Y (2003) Analysis of antioxidative phenolic compounds in artichoke (Cynara scolymus L.). J Agric Food Chem 51(3):601–608
Wang W, Vinocur B, Shoseyov O et al (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9:244–252
Winkel B (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55:85–107
Xu C, Sullivan JH, Garrett WM, Caperna TJ, Natarajan S (2008) Impact of solar ultraviolet-B on the proteome in soybean lines differing in flavonoid contents. Phytochemistry 69(1):38–48
Yamamori A, Onodera S, Kikuchi M et al (2002) Two novel oligosaccharides formed by 1(F)-fructosyltransferase purified from roots of asparagus (Asparagus officinalis L.). Biosci Biotechnol Biochem 66:1419–1422
Yang Q, Wang Y, Zhang J et al (2007) Identification of aluminum-responsive proteins in rice roots by a proteomic approach: cysteine synthase as a key player in Al response. Proteomics 7:737–749
Zhang J, Sun L, Liu L, Lian J, An S, Wang X, Zhang J, Jin J, Li S, Xi J (2010) Proteomic analysis of interactions between the generalist herbivore Spodoptera exigua (Lepidoptera: Noctuidae) and Arabidopsis thaliana. Plant Mol Biol Report 28(2):324–333
Acknowledgements
This research has been enabled by a grant from “Progetto Lagrange-Fondazione CRT”. The research was performed in the context of the project iTECHPLAT Intervention, co-financed by the EU on the Measure 3.4 of the Regione Piemonte docup 2000/2006, supported and implemented by Polo di Innovazione Provinciale of the Province of Turin 2009. Special thanks are due to Franco Fedeli for his training on HPLC analysis.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
Supplementary Material 1
Bar diagram of the UV-C modulated proteins with fold change and standard error. (PPT 232 kb)
Supplementary Material 2
Protein identification from the globe artichoke PEG-enriched proteome 24 h after UV-C irradiation with peptides. (DOC 637 kb)
Rights and permissions
About this article
Cite this article
Falvo, S., Acquadro, A., Albo, A.G. et al. Proteomic Analysis of PEG-Fractionated UV-C Stress-Response Proteins in Globe Artichoke. Plant Mol Biol Rep 30, 111–122 (2012). https://doi.org/10.1007/s11105-011-0325-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-011-0325-2