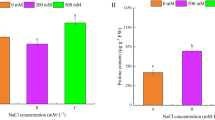
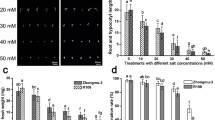
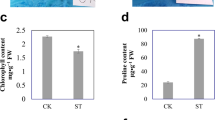
Abstract
Background and aims
Salinity is one of the serious environmental stresses limiting crop growth and yield. We have previously reported that GsCBRLK functions as a positive regulator of plant tolerance to salt stress. In order to investigate the physiological and molecular mechanisms underlying the salinity tolerance regulated by GsCBRLK, this gene was overexpressed in soybean plants. Here we examined the salt-responsive proteomes of the GsCBRLK overexpression soybean and wild type plants using iTRAQ-based proteomic approach to investigate the global effects and potential downstream targets of GsCBRLK.
Results
A total of 941 proteins showed significant changes in protein abundance in soybean leaves, and 574 of the NaCl-regulated proteins were GsCBRLK-dependent. Among the identified proteins, four protein changes in the two genotypes after NaCl treatment were validated using Western blot analysis, at the same time, transcriptional leves of 10 proteins related to Ca2+ signaling were detected using qRT-PCR analysis. Identification of the salt-reponsive proteins has revealed the involvement of GsCBRLK protein in the enhancement of ROS scavenging and photosynthesis capacity in soybean, which was corrobarated with the physiological effects of GsCBRLK overexpression. More importantly, the proteomic data has suggested the regulatory function of GsCBRLK in salt signal transduction pathway mediated by Ca2+/CaM.
Conclusions
These findings have contributed to our knowledge of plant GsCBRLK mediated salt tolerance mechanisms.






Similar content being viewed by others
References
Aghaei K, Ehsanpour AA, Shah AH, Komatsu S (2009) Proteome analysis of soybean hypocotyl and root under salt stress. Amino Acids 36:91–98. doi:10.1007/s00726-008-0036-7
Alvarez S, Hicks LM, Pandey S (2011) ABA-dependent and -independent G-protein signaling in Arabidopsis roots revealed through an iTRAQ proteomics approach. J Proteome Res 10:3107–3122. doi:10.1021/pr2001786
Alvarez S, Roy Choudhury S, Hicks LM, Pandey S (2013) Quantitative proteomics-based analysis supports a significant role of GTG proteins in regulation of ABA response in Arabidopsis roots. J Proteome Res 12:1487–1501. doi:10.1021/pr301159u
Alvarez S, Roy Choudhury S, Sivagnanam K, Hicks LM, Pandey S (2015) Quantitative proteomics analysis of Camelina sativa seeds overexpressing the AGG3 gene to identify the proteomic basis of increased yield and stress tolerance. J Proteome Res 14:2606–2616. doi:10.1021/acs.jproteome.5b00150
Asano T, Hayashi N, Kikuchi S, Ohsugi R (2012) CDPK-mediated abiotic stress signaling. Plant Signal Behav 7:817–821. doi:10.4161/psb.20351
Bai X, Liu J, Tang L, Cai H, Chen M, Ji W, Liu Y, Zhu Y (2013) Overexpression of GsCBRLK from Glycine soja enhances tolerance to salt stress in transgenic alfalfa (Medicago sativa). Funct Plant Biol 40:1048–1056
Bhatt I, Tripathi BN (2011) Plant peroxiredoxins: catalytic mechanisms, functional significance and future perspectives. Biotechnol Adv 29:850–859. doi:10.1016/j.biotechadv.2011.07.002
Bouche N, Yellin A, Snedden WA, Fromm H (2005) Plant-specific calmodulin-binding proteins. Annu Rev Plant Biol 56:435–466. doi:10.1146/annurev.arplant.56.032604.144224
Boudsocq M, Sheen J (2013) CDPKs in immune and stress signaling. Trends Plant Sci 18:30–40. doi:10.1016/j.tplants.2012.08.008
Campo S, Baldrich P, Messeguer J, Lalanne E, Coca M, San Segundo B (2014) Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol 165:688–704. doi:10.1104/pp.113.230268
Cejudo FJ, Meyer AJ, Reichheld JP, Rouhier N, Traverso JA (2014) Thiol-based redox homeostasis and signaling. Front Plant Sci 5:266. doi:10.3389/fpls.2014.00266
Chaves MM, Flexas J, Pinheiro C (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot 103:551–560. doi:10.1093/aob/mcn125
Cho HK, Park JA, Pai HS (2008) Physiological function of NbRanBP1 in Nicotiana benthamiana. Mol Cells 26:270–277
Chung WS, Lee SH, Kim JC, Heo WD, Kim MC, Park CY, Park HC, Lim CO, Kim WB, Harper JF, Cho MJ (2000) Identification of a calmodulin-regulated soybean Ca(2+)-ATPase (SCA1) that is located in the plasma membrane. Plant Cell 12:1393–1407
Dalal M, Tayal D, Chinnusamy V, Bansal KC (2009) Abiotic stress and ABA-inducible Group 4 LEA from Brassica napus plays a key role in salt and drought tolerance. J Biotechnol 139:137–145. doi:10.1016/j.jbiotec.2008.09.014
Dunajska-Ordak K, Skorupa-Klaput M, Kurnik K, Tretyn A, Tyburski J (2014) Cloning and expression analysis of a gene encoding for ascorbate peroxidase and responsive to salt stress in beet (). Plant Mol Biol Report / ISPMB 32:162–175. doi:10.1007/s11105-013-0636-6
Evans C, Noirel J, Ow SY, Salim M, Pereira-Medrano AG, Couto N, Pandhal J, Smith D, Pham TK, Karunakaran E, Zou X, Biggs CA, Wright PC (2012) An insight into iTRAQ: where do we stand now? Anal Bioanal Chem 404:1011–1027. doi:10.1007/s00216-012-5918-6
Hakeem KR, Khan F, Chandna R, Siddiqui TO, Iqbal M (2012) Genotypic variability among soybean genotypes under NaCl stress and proteome analysis of salt-tolerant genotype. Appl Biochem Biotechnol 168:2309–2329
Harmon AC, Gribskov M, Gubrium E, Harper JF (2001) The CDPK superfamily of protein kinases. New Phytol 151:175–183
Harper JE, Breton G, Harmon A (2004) Decoding Ca2+ signals through plant protein kinases. Annu Rev Plant Biol 55:263–288. doi:10.1146/annurev.arplant.55.031903.141627
Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Biol 51:463–499
Hashimoto K, Kudla J (2011) Calcium decoding mechanisms in plants. Biochimie 93:2054–2059. doi:10.1016/j.biochi.2011.05.019
Holmstrom KO, Somersalo S, Mandal A, Palva TE, Welin B (2000) Improved tolerance to salinity and low temperature in transgenic tobacco producing glycine betaine. J Exp Bot 51:177–185
Hou S, Jones SW, Choe LH, Papoutsakis ET, Lee KH (2013) Workflow for quantitative proteomic analysis of Clostridium acetobutylicum ATCC 824 using iTRAQ tags. Methods 61:269–276. doi:10.1016/j.ymeth.2013.03.013
Hua W, Li R-J, Wang L, Lu Y-T (2004) A tobacco calmodulin-binding protein kinase (NtCBK2) induced by high-salt/GA treatment and its expression during floral development and embryogenesis. Plant Sci 166:1253–1259
Huda KMK, Banu MSA, Tuteja R, Tuteja N (2013) Global calcium transducer P-type Ca2 + −ATPases open new avenues for agriculture by regulating stress signalling. J Exp Bot 64:3099–3109
Ji W, Zhu Y, Li Y, Yang L, Zhao X, Cai H, Bai X (2010) Over-expression of a glutathione S-transferase gene, GsGST, from wild soybean (Glycine soja) enhances drought and salt tolerance in transgenic tobacco. Biotechnol Lett 32:1173–1179. doi:10.1007/s10529-010-0269-x
Kao W-Y, Tsai T-T, Tsai H-C, Shih C-N (2006) Response of three Glycine species to salt stress. Environ Exp Bot 56:120–125
Khedr AHA, Abbas MA, Wahid AAA, Quick WP, Abogadallah GM (2003) Proline induces the expression of salt‐stress‐responsive proteins and may improve the adaptation of Pancratium maritimum L. to salt‐stress. J Exp Bot 54:2553–2562
Kim CY, Zhang S (2004) Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. Plant J 38:142–151. doi:10.1111/j.1365-313X.2004.02033.x
Kim WY, Kim CY, Cheong NE, Choi YO, Lee KO, Lee SH, Park JB, Nakano A, Bahk JD, Cho MJ, Lee SY (1999) Characterization of two fungal-elicitor-induced rice cDNAs encoding functional homologues of the rab-specific GDP-dissociation inhibitor. Planta 210:143–149
Kim MC, Chung WS, Yun DJ, Cho MJ (2009) Calcium and calmodulin-mediated regulation of gene expression in plants. Mol Plant 2:13–21. doi:10.1093/mp/ssn091
Knight H, Knight MR (2001) Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci 6:262–267
Koh J, Chen S, Zhu N, Yu F, Soltis PS, Soltis DE (2012) Comparative proteomics of the recently and recurrently formed natural allopolyploid Tragopogon mirus (Asteraceae) and its parents. New Phytol 196:292–305. doi:10.1111/j.1469-8137.2012.04251.x
Komatsu S, Hiraga S, Nouri MZ (2014) Analysis of flooding-responsive proteins localized in the nucleus of soybean root tips. Mol Biol Rep 41:1127–1139. doi:10.1007/s11033-013-2959-7
Kurepa J, Wang S, Li Y, Smalle J (2009) Proteasome regulation, plant growth and stress tolerance. Plant Signal Behav 4:924–927
Laohavisit A, Brown AT, Cicuta P, Davies JM (2010) Annexins: components of the calcium and reactive oxygen signaling network. Plant Physiol 152:1824–1829. doi:10.1104/pp.109.145458
Lee S, Lee EJ, Yang EJ, Lee JE, Park AR, Song WH, Park OK (2004) Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell Online 16:1378–1391
Li XY, Chow CK (1994) An improved method for the measurement of malondialdehyde in biological samples. Lipids 29:73–75
Liu L, Hu X, Song J, Zong X, Li D, Li D (2009) Over-expression of a Zea mays L. protein phosphatase 2C gene (ZmPP2C) in Arabidopsis thaliana decreases tolerance to salt and drought. J Plant Physiol 166:531–542. doi:10.1016/j.jplph.2008.07.008
Liu M, Li D, Wang Z, Meng F, Li Y, Wu X, Teng W, Han Y, Li W (2012) Transgenic expression of ThIPK2 gene in soybean improves stress tolerance, oleic acid content and seed size. Plant Cell Tissue Organ Cult (PCTOC) 111:277–289. doi:10.1007/s11240-012-0192-z
Liu CW, Chang TS, Hsu YK, Wang AZ, Yen HC, Wu YP, Wang CS, Lai CC (2014) Comparative proteomic analysis of early salt stress responsive proteins in roots and leaves of rice. Proteomics 14:1759–1775. doi:10.1002/pmic.201300276
Lv DW, Subburaj S, Cao M, Yan X, Li X, Appels R, Sun DF, Ma W, Yan YM (2014) Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress. Mol Cell Proteomics 13:632–652. doi:10.1074/mcp.M113.030171
Ma X, Qian Q, Zhu D (2005) Expression of a calcineurin gene improves salt stress tolerance in transgenic rice. Plant Mol Biol 58:483–495. doi:10.1007/s11103-005-6162-7
Ma H, Song L, Shu Y, Wang S, Niu J, Wang Z, Yu T, Gu W, Ma H (2012) Comparative proteomic analysis of seedling leaves of different salt tolerant soybean genotypes. J Proteomics 75:1529–1546. doi:10.1016/j.jprot.2011.11.026
Manaa A, Faurobert M, Valot B, Bouchet J-P, Grasselly D, Causse M, Ahmed HB (2013) Effect of salinity and calcium on tomato fruit proteome. OMICS 17:338–352
Murad AM, Molinari HB, Magalhaes BS, Franco AC, Takahashi FS, de Oliveira NG, Franco OL, Quirino BF (2014) Physiological and proteomic analyses of Saccharum spp. grown under salt stress. PLoS One 9, e98463. doi:10.1371/journal.pone.0098463
Nguyen TH, Brechenmacher L, Aldrich JT, Clauss TR, Gritsenko MA, Hixson KK, Libault M, Tanaka K, Yang F, Yao Q, Pasa-Tolic L, Xu D, Nguyen HT, Stacey G (2012) Quantitative phosphoproteomic analysis of soybean root hairs inoculated with Bradyrhizobium japonicum. Mol Cell Proteomics 11:1140–1155. doi:10.1074/mcp.M112.018028
Pandey S, Tiwari SB, Tyagi W, Reddy MK, Upadhyaya KC, Sopory SK (2002) A Ca2+/CaM‐dependent kinase from pea is stress regulated and in vitro phosphorylates a protein that binds to AtCaM5 promoter. Eur J Biochem 269:3193–3204
Parker J, Balmant K, Zhu F, Zhu N, Chen S (2015) cysTMTRAQ-An integrative method for unbiased thiol-based redox proteomics. Mol Cell Proteomics 14:237–242. doi:10.1074/mcp.O114.041772
Pi E, Qu L, Hu J, Huang Y, Qiu L, Lu H, Jiang B, Liu C, Peng T, Zhao Y, Wang H, Tsai SN, Ngai S, Du L (2015) Mechanisms of soybean roots’ tolerances to salinity revealed by proteomic and phosphoproteomic comparisons between two cultivars. Mol Cell Proteomics. doi:10.1074/mcp.M115.051961
Pruzinska A, Anders I, Aubry S, Schenk N, Tapernoux-Luthi E, Muller T, Krautler B, Hortensteiner S (2007) In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19:369–387. doi:10.1105/tpc.106.044404
Putz SM, Boehm AM, Stiewe T, Sickmann A (2012) iTRAQ analysis of a cell culture model for malignant transformation, including comparison with 2D-PAGE and SILAC. J Proteome Res 11:2140–2153. doi:10.1021/pr200881c
Qin J, Gu F, Liu D, Yin C, Zhao S, Chen H, Zhang J, Yang C, Zhan X, Zhang M (2013) Proteomic analysis of elite soybean Jidou17 and its parents using iTRAQ-based quantitative approaches. Proteome Sci 11:12. doi:10.1186/1477-5956-11-12
Qudeimat E, Frank W (2009) Ca2+ signatures: the role of Ca2 + −ATPases. Plant Signal Behav 4:350–352
Qudeimat E, Faltusz AM, Wheeler G, Lang D, Holtorf H, Brownlee C, Reski R, Frank W (2008) A PIIB-type Ca2 + −ATPase is essential for stress adaptation in Physcomitrella patens. Proc Natl Acad Sci 105:19555–19560
Ranty B, Aldon D, Galaud JP (2006) Plant calmodulins and calmodulin-related proteins: multifaceted relays to decode calcium signals. Plant Signal Behav 1:96–104
Richards SL, Laohavisit A, Mortimer JC, Shabala L, Swarbreck SM, Shabala S, Davies JM (2014) Annexin 1 regulates the H2O2-induced calcium signature in Arabidopsis thaliana roots. Plant J 77:136–145. doi:10.1111/tpj.12372
Roberts MR, Salinas J, Collinge DB (2002) 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol Biol 50:1031–1039
Romano PG, Horton P, Gray JE (2004) The Arabidopsis cyclophilin gene family. Plant Physiol 134:1268–1282. doi:10.1104/pp.103.022160
Rusnak F, Mertz P (2000) Calcineurin: form and function. Physiol Rev 80:1483–1521
Singh DK, McNellis TW (2011) Fibrillin protein function: the tip of the iceberg? Trends Plant Sci 16:432–441. doi:10.1016/j.tplants.2011.03.014
Snedden WA, Fromm H (2001) Calmodulin as a versatile calcium signal transducer in plants. New Phytol 151:35–66
Sobhanian H, Razavizadeh R, Nanjo Y, Ehsanpour AA, Jazii FR, Motamed N, Komatsu S (2010) Proteome analysis of soybean leaves, hypocotyls and roots under salt stress. Proteome Sci 8:19. doi:10.1186/1477-5956-8-19
Sobhanian H, Aghaei K, Komatsu S (2011) Changes in the plant proteome resulting from salt stress: toward the creation of salt-tolerant crops? J Proteomics 74:1323–1337. doi:10.1016/j.jprot.2011.03.018
Suo J, Zhao Q, Zhang Z, Chen S, Cao J, Liu G, Wei X, Wang T, Yang C, Dai S (2015) Cytological and proteomic analyses of Osmunda cinnamomea germinating spores reveal characteristics of fern spore germination and rhizoid tip growth. Mol Cell Proteomics 14:2510–2534. doi:10.1074/mcp.M114.047225
Tester M, Langridge P (2010) Breeding technologies to increase crop production in a changing world. Science 327:818–822. doi:10.1126/science.1183700
Tran LS, Mochida K (2010) Functional genomics of soybean for improvement of productivity in adverse conditions. Funct Integr Genom 10:447–462. doi:10.1007/s10142-010-0178-z
Tuteja N (2007) Mechanisms of high salinity tolerance in plants. Methods Enzymol 428:419–438. doi:10.1016/S0076-6879(07)28024-3
Vemuri GN, Aristidou AA (2005) Metabolic engineering in the -omics era: elucidating and modulating regulatory networks. Microbiol Mol Biol Rev 69:197–216. doi:10.1128/MMBR.69.2.197-216.2005
Vera-Estrella R, Barkla BJ, Garcia-Ramirez L, Pantoja O (2005) Salt stress in Thellungiella halophila activates Na + transport mechanisms required for salinity tolerance. Plant Physiol 139:1507–1517. doi:10.1104/pp.105.067850
Wang X, Chen S, Zhang H, Shi L, Cao F, Guo L, Xie Y, Wang T, Yan X, Dai S (2010a) Desiccation tolerance mechanism in resurrection fern-ally Selaginella tamariscina revealed by physiological and proteomic analysis. J Proteome Res 9:6561–6577. doi:10.1021/pr100767k
Wang YC, Qu GZ, Li HY, Wu YJ, Wang C, Liu GF, Yang CP (2010b) Enhanced salt tolerance of transgenic poplar plants expressing a manganese superoxide dismutase from Tamarix androssowii. Mol Biol Rep 37:1119–1124. doi:10.1007/s11033-009-9884-9
Wang P, Li Z, Wei J, Zhao Z, Sun D, Cui S (2012) A Na+/Ca2+ exchanger-like protein (AtNCL) involved in salt stress in Arabidopsis. J Biol Chem 287:44062–44070. doi:10.1074/jbc.M112.351643
Wang G, Zeng H, Hu X, Zhu Y, Chen Y, Shen C, Wang H, Poovaiah BW, Du L (2014a) Identification and expression analyses of calmodulin-binding transcription activator genes in soybean. Plant Soil 386:205–221. doi:10.1007/s11104-014-2267-6
Wang L, Liu X, Liang M, Tan F, Liang W, Chen Y, Lin Y, Huang L, Xing J, Chen W (2014b) Proteomic analysis of salt-responsive proteins in the leaves of mangrove Kandelia candel during short-term stress. PLoS One 9, e83141
Watillon B, Kettmann R, Boxus P, Burny A (1993) A calcium/calmodulin-binding serine/threonine protein kinase homologous to the mammalian type II calcium/calmodulin-dependent protein kinase is expressed in plant cells. Plant Physiol 101:1381–1384
Weinl S, Kudla J (2009) The CBL-CIPK Ca(2+)-decoding signaling network: function and perspectives. New Phytol 184:517–528
Xiao X, Ma J, Wang J, Wu X, Li P, Yao Y (2014) Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Front Plant Sci 5:788. doi:10.3389/fpls.2014.00788
Xu S (2010) Abscisic acid activates a Ca2 + −calmodulin-stimulated protein kinase involved in antioxidant defense in maize leaves. Acta Biochim Biophys Sin (Shanghai) 42:646–655. doi:10.1093/abbs/gmq064
Yamazaki T, Kawamura Y, Minami A, Uemura M (2008) Calcium-dependent freezing tolerance in Arabidopsis involves membrane resealing via synaptotagmin SYT1. Plant Cell 20:3389–3404. doi:10.1105/tpc.108.062679
Yang T, Poovaiah BW (2003) Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci 8:505–512. doi:10.1016/j.tplants.2003.09.004
Yang T, Chaudhuri S, Yang L, Chen Y, Poovaiah B (2004) Calcium/calmodulin up-regulates a cytoplasmic receptor-like kinase in plants. J Biol Chem 279:42552–42559
Yang L, Ji W, Zhu Y, Gao P, Li Y, Cai H, Bai X, Guo D (2010a) GsCBRLK, a calcium/calmodulin-binding receptor-like kinase, is a positive regulator of plant tolerance to salt and ABA stress. J Exp Bot 61:2519–2533. doi:10.1093/jxb/erq084
Yang T, Chaudhuri S, Yang L, Du L, Poovaiah BW (2010b) A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J Biol Chem 285:7119–7126. doi:10.1074/jbc.M109.035659
Yang C, Li A, Zhao Y, Zhang Z, Zhu Y, Tan X, Geng S, Guo H, Zhang X, Kang Z (2011) Overexpression of a wheat CCaMK gene reduces ABA sensitivity of Arabidopsis thaliana during seed germination and seedling growth. Plant Mol Biol Rep 29:681–692
Yang L, Zhang Y, Zhu N, Koh J, Ma C, Pan Y, Yu B, Chen S, Li H (2013a) Proteomic analysis of salt tolerance in sugar beet monosomic addition line M14. J Proteome Res 12:4931–4950. doi:10.1021/pr400177m
Yang LT, Qi YP, Lu YB, Guo P, Sang W, Feng H, Zhang HX, Chen LS (2013b) iTRAQ protein profile analysis of Citrus sinensis roots in response to long-term boron-deficiency. J Proteomics 93:179–206. doi:10.1016/j.jprot.2013.04.025
Yazaki K (2006) ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett 580:1183–1191. doi:10.1016/j.febslet.2005.12.009
Zhang CJ, Zhao BC, Ge WN, Zhang YF, Song Y, Sun DY, Guo Y (2011) An apoplastic h-type thioredoxin is involved in the stress response through regulation of the apoplastic reactive oxygen species in rice. Plant Physiol 157:1884–1899. doi:10.1104/pp.111.182808
Zhang H, Han B, Wang T, Chen S, Li H, Zhang Y, Dai S (2012) Mechanisms of plant salt response: insights from proteomics. J Proteome Res 11:49–67. doi:10.1021/pr200861w
Zhu JK (2001a) Cell signaling under salt, water and cold stresses. Curr Opin Plant Biol 4:401–406
Zhu JK (2001b) Plant salt tolerance. Trends Plant Sci 6:66–71
Zhu MM, Dai SJ, McClung S, Yan XF, Chen SX (2009) Functional differentiation of Brassica napus guard cells and mesophyll cells revealed by comparative proteomics. Mol Cell Proteomics 8:752–766. doi:10.1074/mcp.M800343-MCP200
Zhu M, Dai S, Zhu N, Booy A, Simons B, Yi S, Chen S (2012) Methyl jasmonate responsive proteins in Brassica napus guard cells revealed by iTRAQ-based quantitative proteomics. J Proteome Res 11:3728–3742. doi:10.1021/pr300213k
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31401311), China Postdoctoral Science Foundation (2013T60341, 2012M510915), Doctoral Fund of Ministry of Education of China (20112325120020) and Harbin municipal science and technology research fund innovative talents project (2014RFQXJ002).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Frans J.M Maathuis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table 1
Primer list for qRT-PCR of differential expressed proteins identified from iTRAQ (XLSX 160 kb)
Supplemental Table 2
List of the 2118 differentially expressed proteins identified in soybean leaves after merging the 3 replicates. (XLSX 521 kb)
Supplemental Table 3
List of proteins that respond to NaCl in soybean leaves of WT (115/113) and/or OE#9 (116/114)a (XLSX 244 kb)
Supplemental Table 4
List of proteins that respond to NaCl only in soybean leaves of GsCBRLK overexpression line OE#9 (116/114)a (XLSX 213 kb)
Supplemental Table 5
Proteins affected by overexpression of GsCBRLK protein in the absence of NaCl (114/113)a (XLSX 551 kb)
Supplemental Fig. 1
Map of the plasmid construct. The GsCBRLK gene was under the control of the CaMV 35S promoter with the binding enhancer E12. The bar gene was used as the selective marker gene. LB the left T-DNA border, and RB the right T-DNA border. (PPTX 56 kb)
Supplemental Fig. 2
Schematics of the experimental design for the iTRAQ proteomic analysis. WT, wild-type soybean plants; OE#9, GsCBRLK overexpression plants . (PPTX 120 kb)
Supplemental Fig. 3
Evaluation of GsCBRLK expression levels in transgenic soybean lines (A) RT–PCR analysis of GsCBRLK expression in leaves of control and transgenic soybean. The PCR products of 200 bp were resolved on 2.0 % agarose gel. WT, non-transformed (wild-type) plants; OE#9, OE#11, transgenic plants overexpressing GsCBRLK. A 194 bp 18SrRNA gene fragment was amplified by RT–PCR as an internal control. (B) Western blot analysis of GsCBRLK expression in leaves of control and transgenic soybean. Coomassie stained gel image showing equal loading of protein samples (10 μg each lane) from WT and transgenic plants OE#9 and OE#11. (PPTX 135 kb)
Supplemental Fig. 4
Genomic PCR analysis confirming stably inherited of GsCBRLK in transgenic soybean plants. The characteristic 750 bp fragment of the Bar gene was amplified. +, plasmid pCEOM-GsCBRLK positive control; −, ddH2O negative control; WT, wild-type untransformed soybean control. (A) PCR detection of T0 transgenic lines, 1–12, independent transgenic soybean lines; (B) PCR detection of transgenic soybean T1 plants, 4 independent plants from transgenic lines OE#9, OE#10 and OE#11 seperately; (C) PCR detection of transgenic soybean T2 plants, 3 independent plants from T1 plants of transgenic lines OE#9 and OE#11 seperately. (PPTX 250 kb)
Supplemental Fig. 5
Functional classification for the 2118 differentially expressed proteins in soybean leaves. (PPTX 78 kb)
Rights and permissions
About this article
Cite this article
Ji, W., Koh, J., Li, S. et al. Quantitative proteomics reveals an important role of GsCBRLK in salt stress response of soybean. Plant Soil 402, 159–178 (2016). https://doi.org/10.1007/s11104-015-2782-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-015-2782-0