Abstract
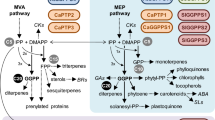
Geranylgeranyl diphosphate (GGPP) is a key precursor of various isoprenoids that have diverse functions in plant metabolism and development. The annotation of the Arabidopsis thaliana genome predicts 12 genes to encode geranylgeranyl diphosphate synthases (GGPPS). In this study we analyzed GGPPS activity as well as the subcellular localization and tissue-specific expression of the entire protein family in A. thaliana. GGPPS2 (At2g18620), GGPPS3 (At2g18640), GGPPS6 (At3g14530), GGPPS7 (At3g14550), GGPPS8 (At3g20160), GGPPS9 (At3g29430), GGPPS10 (At3g32040) and GGPPS11 (At4g36810) showed GGPPS activity in Escherichia coli, similar to activities reported earlier for GGPPS1 (At1g49530) and GGPPS4 (At2g23800) (Zhu et al. in Plant Cell Physiol 38(3):357–361, 1997a; Plant Mol Biol 35(3):331–341, b). GGPPS12 (At4g38460) did not produce GGPP in E. coli. Based on DNA sequence analysis we propose that GGPPS5 (At3g14510) is a pseudogene. GGPPS–GFP (green fluorescent protein) fusion proteins of the ten functional GGPP synthases localized to plastids, mitochondria and the endoplasmic reticulum, with the majority of the enzymes located in plastids. Gene expression analysis using quantitative real time-PCR, GGPPS promoter-GUS (β-glucuronidase) assays and publicly available microarray data revealed a differential spatio-temporal expression of GGPPS genes. The results suggest that plastids and mitochondria are key subcellular compartments for the synthesis of ubiquitous GGPP-derived isoprenoid species. GGPPS11 and GGPPS1 are the major isozymes responsible for their biosynthesis. All remaining paralogs, encoding six plastidial isozymes and two cytosolic isozymes, were expressed in specific tissues and/or at specific developmental stages, suggesting their role in developmentally regulated isoprenoid biosynthesis. Our results show that of the 12 predicted GGPPS encoded in the A. thaliana genome 10 are functional proteins that can synthesize GGPP. Their specific subcellular location and differential expression pattern suggest subfunctionalization in providing GGPP to specific tissues, developmental stages, or metabolic pathways.












Similar content being viewed by others
Abbreviations
- ABA:
-
Abscisic acid
- DMAPP:
-
Dimethylallyl diphosphate
- ER:
-
Endoplasmic reticulum
- FPP:
-
Farnesyl diphosphate
- GA:
-
Gibberellic acid
- GFP:
-
Green fluorescent protein
- GGPP:
-
Geranylgeranyl diphosphate
- GGPPS:
-
Geranylgeranyl diphosphate synthase
- GPP:
-
Geranyl diphosphate
- GUS:
-
β-Glucuronidase
- IPP:
-
Isopentenyl diphosphate
- MEP:
-
Methylerythritol
- MVA:
-
Mevalonate
References
Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol 16(6):553–563
Bick JA, Lange BM (2003) Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Arch Biochem Biophys 415(2):146–154
Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302(5652):1956–1960
Bouvier F, Suire C, d’Harlingue A, Backhaus RA, Camara B (2000) Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. Plant J 24(2):241–252
Caughey WS, Smythe GA, O’Keeffe DH, Maskasky JE, Smith MI (1975) Heme A of cytochrome c oxicase. Structure and properties: comparisons with hemes B, C, and S and derivatives. J Biol Chem 250(19):7602–7622
Closa M, Vranová E, Bortolotti C, Bigler L, Arró M, Ferrer A, Gruissem W (2010) The Arabidopsis thaliana FPP synthase isozymes have overlapping and specific functions in isoprenoid biosynthesis, and complete loss of FPP synthase activity causes early developmental arrest. Plant J 63(3):512–525
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743
Cunillera N, Arró M, Delourme D, Karst F, Boronat A, Ferrer A (1996) Arabidopsis thaliana contains two differentially expressed farnesyl-diphosphate synthase genes. J Biol Chem 271(13):7774–7780
Cunillera N, Boronat A, Ferrer A (1997) The Arabidopsis thaliana FPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate synthase isoform. J Biol Chem 272(24):15381–15388
Czechowski T, Bari RP, Stitt M, Scheible W-R, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38(2):366–379
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139(1):5–17
Delourme D, Lacroute F, Karst F (1994) Cloning of an Arabidopsis thaliana cDNA coding for farnesyl diphosphate synthase by functional complementation in yeast. Plant Mol Biol 26(6):1867–1873
Ducluzeau A-L, Wamboldt Y, Elowsky CG, Mackenzie SA, Schuurink RC, Basset GJC (2011) Gene network reconstruction identifies the authentic trans-prenyl diphosphate synthase that makes the solanesyl moiety of ubiquinone-9 in Arabidopsis. Plant J 69(2):366–375
Dugardeyn J, Vandenbussche F, Van Der Straeten D (2008) To grow or not to grow: what can we learn on ethylene-gibberellin cross-talk by in silico gene expression analysis? J Exp Bot 59(1):1–16
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300(4):1005–1016
Floss DS, Walter M (2009) Role of carotenoid cleavage dioxygenase 1(CCD1) in apocarotenoid biogenesis revisited. Plant Signaling Behav 4(3):172–175
Flügge U-I, Gao W (2005) Transport of isoprenoid intermediates across chloroplast envelope membranes. Plant Biol 1:91–97
Hojo M, Matsumoto T, Miura T (2007) Cloning and expression of a geranylgeranyl diphosphate synthase gene: insights into the synthesis of termite defence secretion. Insect Mol Biol 16(1):121–131
Hruz T, Wyss M, Docquier M, Pfaffl M, Masanetz S, Borghi L, Verbrugghe P, Kalaydjieva L, Bleuler S, Laule O, Descombes P, Gruissem W, Zimmermann P (2011) RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genomics 12(1):156
Hsieh F-L, Chang T-H, Ko T-P, Wang AH-J (2011) Structure and mechanism of an Arabidopsis medium/long-chain-length prenyl pyrophosphate synthase. Plant Physiol 155(3):1079–1090
Hu J, Mitchum MG, Barnaby N, Ayele BT, Ogawa M, Nam E, Lai W-C, Hanada A, Alonso JM, Ecker JR, Swain SM, Yamaguchi S, Kamiya Y, Sun T-p (2008) Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell 20(2):320–336
Huang M, Abel C, Sohrabi R, Petri J, Haupt I, Cosimano J, Gershenzon J, Tholl D (2010) Variation of herbivore-induced volatile terpenes among Arabidopsis ecotypes depends on allelic differences and subcellular targeting of two terpene synthases, TPS02 and TPS03. Plant Physiol 153(3):1293–1310
Jiang Y, Proteau P, Poulter D, Ferro-Novick S (1995) BTS1 encodes a geranylgeranyl diphosphate synthase in Saccharomyces cerevisiae. J Biol Chem 270(37):21793–21799
Joyard J, Ferro M, Masselon C, Seigneurin-Berny D, Salvi D, Garin Jrm, Rolland N (2009) Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol Plant 2(6):1154–1180
Kainou T, Kawamura K, Tanaka K, Matsuda H, Kawamukai M (1999) Identification of the GGPS1 genes encoding geranylgeranyl diphosphate synthases from mouse and human. Biochim Biophys Acta Mol Cell Biol Lipids 1437(3):333–340
Karimi M, De DeMeyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10(3):103–105
Karlen Y, McNair A, Perseguers S, Mazza C, Mermod N (2007) Statistical significance of quantitative PCR. BMC Bioinformatics 8(1):131
Kellogg BA, Poulter CD (1997) Chain elongation in the isoprenoid biosynthetic pathway. Curr Opin Chem Biol 1(4):570–578
Koncz C, Shell C (1986) The promoter of T1-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by novel type of Agrobacterium binary vector. Mol Gen Genet 204:383–396
Kozak M (1997) Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J 16(9):2482–2492
Kuepfer L, Sauer U, Blank LM (2005) Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res 15(10):1421–1430
Lange BM, Ghassemian M (2003) Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol 51(6):925–948
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, Drews GN, Fischer RL, Okamuro JK, Harada JJ, Goldberg RB (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107(18):8063–8070
Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A (2006) Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J 45(3):309–319
Massonnet C, Vile D, Fabre J, Hannah MA, Caldana C, Lisec J, Beemster GT, Meyer RC, Messerli G, Gronlund JT, Perkovic J, Wigmore E, May S, Bevan MW, Meyer C, Rubio-Diaz S, Weigel D, Micol JL, Buchanan-Wollaston V, Fiorani F, Walsh S, Rinn B, Gruissem W, Hilson P, Hennig L, Willmitzer L, Granier C (2010) Probing the reproducibility of leaf growth and molecular phenotypes: a comparison of three Arabidopsis accessions cultivated in ten laboratories. Plant Physiol 152(4):2142–2157
Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K (1990) Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol 172(12):6704–6712
Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun T-p (2006) Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J 45(5):804–818
Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56(1):165–185
Nelson BK, Cai X, Nebenfuhr A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51(6):1126–1136
Nishino T, Rudney H (1977) Effects of detergents on the properties of 4-hydroxybenzoate. Polyprenyl transferase and the specificity of the polyprenyl pyrophosphate synthetic system in mitochondria. Biochemistry 16(4):605–609
Ohnuma S, Koyama T, Ogura K (1993) Alteration of the product specificities of prenyltransferases by metal ions. Biochem Bioph Res Co 192(2):407–412
Ohnuma S, Suzuki M, Nishino T (1994) Archaebacterial ether-linked lipid biosynthetic gene. Expression cloning, sequencing, and characterization of geranylgeranyl-diphosphate synthase. J Biol Chem 269(20):14792–14797
Okada K, Saito T, Nakagawa T, Kawamukai M, Kamiya Y (2000) Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol 122(4):1045–1056
Pan J–J, Kuo T-H, Chen Y-K, Yang L-W, Po-Huang L (2002) Insight into the activation mechanism of Escherichia coli octaprenyl pyrophosphate synthase derived from pre-steady-state kinetic analysis. Biochim Biophys Acta Protein Struct Mol Enzymol 1594(1):64–73
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45
Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K (2009) PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21(12):3718–3731
Rangan L, Vogel C, Srivastava A (2008) Analysis of context sequence surrounding translation initiation site from complete genome of model plants. Mol Biotechnol 39(3):207–213
Rieu I, Powers SJ (2009) Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell 21(4):1031–1033
Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37(6):e45
Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, Verstappen F, Bouwmeester H (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol 155(2):721–734
Sandmann G, Misawa N, Wiedemann M, Vittorioso P, Carattoli A, Morelli G, Macino G (1993) Functional identification of al-3 from Neurospora crassa as the gene for geranylgeranyl pyrophosphate synthase by complementation with crt genes, in vitro characterization of the gene product and mutant analysis. J Photoch Photobio B 18(2–3):245–251
Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37(5):501–506
Smyth D, Bowman J, Meyerowitz E (1990) Early flower development in Arabidopsis. Plant Cell 2:755–767
Tan B-C, Joseph LM, Deng W-T, Liu L, Li Q-B, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35(1):44–56
Tholl D, Lee S (2011) Terpene Specialized Metabolism in Arabidopsis thaliana. The Arabidopsis Book:e0143
Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The botany array resource: e-Northerns, expression angling, and promoter analyses. Plant J 43(1):153–163
Ubeda-Tomas S, Swarup R, Coates J, Swarup K, Laplaze L, Beemster GTS, Hedden P, Bhalerao R, Bennett MJ (2008) Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol 10(5):625–628
Ubeda-Tomás S, Federici F, Casimiro I, Beemster GTS, Bhalerao R, Swarup R, Doerner P, Haseloff J, Bennett MJ (2009) Gibberellin signaling in the endodermis controls arabidopsis root meristem size. Curr Biol 19(14):1194–1199
Vallon T, Ghanegaonkar S, Vielhauer O, Muller A, Albermann C, Sprenger G, Reuss M, Lemuth K (2008) Quantitative analysis of isoprenoid diphosphate intermediates in recombinant and wild-type Escherichia coli strains. Appl Microbiol Biotechnol 81(1):175–182
van Schie CC, Ament K, Schmidt A, Lange T, Haring MA, Schuurink RC (2007) Geranyl diphosphate synthase is required for biosynthesis of gibberellins. Plant J 52(4):752–762
Vandermoten S, Haubruge E, Cusson M (2009) New insights into short-chain prenyltransferases: structural features, evolutionary history and potential for selective inhibition. Cell Mol Life Sci 66(23):3685–3695
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):research0034.0031–research0034.0011
Vranová E, Hirsch-Hoffmann M, Gruissem W (2011) AtIPD: a curated database of arabidopsis isoprenoid pathway models and genes for isoprenoid network analysis. Plant Physiol 156(4):1655–1660
Wang G, Dixon RA (2009) Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc Natl Acad Sci USA 106(24):9914–9919
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59(1):225–251
Zhang J (2003) Evolution by gene duplication: an update. Trends Ecol Evol 18(6):292–298
Zhu X, Suzuki K, Okada K, Tanaka K, Nakagawa T, Kawamukai M, Matsuda K (1997a) Cloning and functional expression of a novel geranylgeranyl pyrophosphate synthase gene from Arabidopsis thaliana in Escherichia coli. Plant Cell Physiol 38(3):357–361
Zhu X, Suzuki K, Saito T, Okada K, Tanaka K, Nakagawa T, Matsuda H, Kawamukai M (1997b) Geranylgeranyl pyrophosphate synthase encoded by the newly isolated gene GGPS6 from Arabidopsis thaliana is localized in mitochondria. Plant Mol Biol 35(3):331–341
Acknowledgments
This work was supported by a grant from ETH Zurich (TH-51 06-1) and the EU FP7 contract 245143 (TiMet). The Spanish Ministerio de Ciencia e Innovacion (www.micinn.es) provided grants BIO2008-00432 and BIO2011-23680 to MRC and a doctoral FPI fellowship to ARS. We thank Biswapriya Biswavas Misra and Christian Barucker for their contribution to the work on subcellular localization. We thank Dr. Axel Schmidt for useful discussions on enzymatic activity assays.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
The gene ID numbers of the GGPPS characterized in this study are: GGPPS1 (GGPPS6 in Zhu et al. 1997b; Okada et al. 2000) is At1g49530; GGPPS2 is At2g18620; GGPPS3 (GGPPS4 in Okada et al. 2000) is At2g18640; GGPPS4 (GGPPS5 in Zhu et al. 1997a; GGPPS2 in Okada et al. 2000) is At2g23800; GGPPS5 is At3g14510; GGPPS6 is At3g14530; GGPPS7 (GGPPS3 in Okada et al. 2000) is At3g14550; GGPPS8 is At3g20160; GGPPS9 is At3g29430; GGPPS10 is At3g32040; GGPPS11 (GGPPS1 in Okada et al. 2000) is At4g36810; GGPPS12 (GGR in Okada et al. 2000) is At4g38640.
Gilles Beck, Diana Coman contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2013_70_MOESM2_ESM.pdf
Supplemental Table S2 as PDF Oligonucleotides used for GGPPSs amplification to construct pENTR/D-TOPO-GGPPS-3` (PDF 235 kb)
11103_2013_70_MOESM4_ESM.pdf
Supplemental Table S4 as PDF Oligonucleotides used for GGPPSs promoter amplification to construct pENTR-D-TOPO-GGPPSpro (PDF 34 kb)
Rights and permissions
About this article
Cite this article
Beck, G., Coman, D., Herren, E. et al. Characterization of the GGPP synthase gene family in Arabidopsis thaliana . Plant Mol Biol 82, 393–416 (2013). https://doi.org/10.1007/s11103-013-0070-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-013-0070-z