Abstract
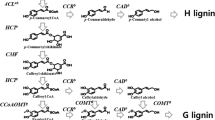
The isolation and characterization of cDNA and homologous genomic clones encoding the lignin O-methyltransferase (OMT) from maize is reported. The cDNA clone has been isolated by differential screening of maize root cDNA library. Southern analysis indicates that a single gene codes for this protein. The genomic sequence contains a single 916 bp intron. The deduced protein sequence from DNA shares significant homology with the recently reported lignin-bispecific caffeic acid/5-hydroxyferulic OMTs from alfalfa and aspen. It also shares homology with OMTs from bovine pineal glands and a purple non-sulfur photosynthetic bacterium. The mRNA of this gene is present at different levels in distinct organs of the plant with the highest accumulation detected in the elongation zone of roots. Bacterial extracts from clones containing the maize OMT cDNA show an activity in methylation of caffeic acid to ferulic acid comparable to that existing in the plant extracts. These results indicate that the described gene encodes the caffeic acid 3-O-methyltransferase (COMT) involved in the lignin biosynthesis of maize.
Similar content being viewed by others
References
Armstrong GA, Alberti M, Leach F, Hearst JE: Nucleotide sequence, organization, and nature of the protein products of the carotenoid biosynthesis gene cluster of Rhodobacter capsulatus. Mol Gen Genet 216: 254–268 (1989).
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K: Current Protocols in Molecular Biology. Greene Publishing Associates/Wiley-Interscience, New York (1990).
Bertocci B, Miggiano V, DaPrada M, Dembic Z, Lahm H-W, Malherbe P: Human catechol-O-methyltransferase: Cloning and expression of the membrane-associated. Proc Natl Acad Sci USA 88: 1416–1420 (1991).
Brown SA: Coumarins. In: Stumpf PK, Conn EE (eds) The Biochemistry of Plants: A Comprehensive Treatise, vol. 7: Secondary Plant Products, pp. 269–300. Academic Press, New York (1981).
Bugos RC, Chiang VL, Campbell WH: Isolation of O-methyltransferase associated with lignin biosynthesis in aspen. Annual Meeting of the American Society of Plant Physiologists, Indianapolis, Indiana. Plant Physiol 93: 15 (1990).
Bugos RC, Chiang VL, Campbell WH: cDNA cloning, sequence analysis and seasonal expression of ligninbispecific caffeic acid/5-hydroxyferulic acid O-methyltransferase of aspen. Plant Mol Biol 17: 1203–1215 (1991).
Campbell WH, Gowri G: Codon usage in higher plants, green algae, and cyanobacteria. Plant Physiol 92: 1–11 (1990).
Charriere-Ladreix Y: O-methyltransferases endoplasmiques de Populus nigra. Phytochemistry 18: 43–45 (1979).
Collendavelloo J, Legrand M, Fritig B: Plant disease and the regulation of enzymes involved in lignification. Plant Physiol 73: 550–554 (1983).
Dalkin K, Edwards R, Edington B, Dixon RA: Stress responses in alfalfa (Medicago sativa L.). Plant Physiol 92: 440–446 (1990).
DeCarolis E, Ibrahim RK: Purification and kinetics of phenylpropanoid O-methyltransferase activities from Brassica oleracea. Biochem Cell Biol 67 763–769 (1989).
Dessen P, Fondrat C, Valencien C, Mugnier C. BISANCE: A French service for access to biomolecular sequence databases. CABIOS 6: 355–356 (1990).
Dumas B, VanDoorsselaere J, Gielen J, Legrand M, Fritig B, VanMontagu M, Inzé D: Nucleotide sequence of a complementary DNA encoding O-methyltransferase from poplar. Plant Physiol 98: 796–797 (1992).
Ebel J, Hahlbrock K, Grisebach H: Purification and properties of an O-dihydriophenol meta-O-methyltransferase from cell suspension cultures of parsley and its relation to flavonoid biosynthesis. Biochim Biophys Acta 268: 313–326 (1972).
Edwards R, Dixon RA: Purification and characterization of S-adenosyl-L-methionine: caffeic acid 3-O-methyltransferase from suspension cultures of alfalfa (Medicago sativa L.). Arch Biochem Biophys 287: 372–379 (1991).
Finkle BJ, Kelly SH: Catechol O-methyltransferases in pampas grass: differentiation of m- and p-methylating activities. Phytochemistry 13: 1719–1725 (1974).
Finkle BJ, Masri MS: Methylation of polyhydroxyaromatic compounds by pampas grass O-methyltransferase. Biochim Biophys Acta 85: 167–169 (1964).
Finkle BJ, Nelson RF: Enzyme reactions with phenolic compounds: A meta-O-methyltransferase in plants. Biochim Biophys Acta 76: 747–749 (1963).
Fukuda H, Komamine A: Lignin synthesis and its related enzymes as markers of tracheary-element differentiation in single cells isolated from the mesophyll of Zinnia elegans. Planta 155: 423–430 (1982).
Gowri G, Bugos RC, Campbell WH, Maxwell CA, Dixon RA: Stress responses in alfalfa (Medicago sativa L.). Plant Physiol 97: 7–14 (1991).
Grisebach H: Lignins. In: Stumpf PK, Conn EE (eds) The Biochemistry of Plants: A Comprehensive Treatise, vol. 7: Secondary Plant Products, pp. 457–478. Academic Press, New York (1981).
Hahlbrock K: Flavonoids. In: Stumpf PK, Conn EE (eds) The Biochemistry of Plants: A Comprehensive Treatise, vol. 7: Secondary Plant Products, pp. 425–456. Academic Press, New York (1981).
Hanahan D: Techniques for transformation of E. coli. In: Glover DM (eds.) DNA Cloning: A Practical Approach, pp. 109–137. IRL Press, Oxford (1985).
Hermann C, Legrand M, Geoffroy P, Fritig B: Enzymatic synthesis of lignin: purification to homogeneity of the three O-methyltransferases of tobacco and production of specific antibodies. Arch Biochem Biophys 253: 367–376 (1987).
Ingrosso D, Fowler AV, Bleibaum J, Clarke S: Sequence of the D-aspartyl/L-isoaspartyl protein methyltransferase from human erythrocytes. J Biol Chem 264: 20131–20139 (1989).
Ishida I, Obinata M, Deguchi T: Molecular cloning and nucleotide sequence of cDNA encoding hydroxyindole O-methyltransferase of bovine pineal glands. J Biol Chem 262: 2895–2899 (1987).
Kuroda H: Comparative studies on O-methyltransferases involved in lignin biosynthesis. Wood Res 69: 91–135 (1983).
Kuroda H, Higuchi T: Characterization and biosynthesis of mistletoe lignin. Phytochemistry 15: 1511–1514 (1976).
Kuroda H, Shimada M, Higuchi T: Purification and properties of O-methyltransferase involved in the biosynthesis of gymnosperm lignin. Phytochemistry 14: 1759–1763 (1975).
Kuroda H, Shimada M, Higuchi T: Characterization of a lignin-specific O-methyltransferase in aspen wood. Phytochemistry 20: 2635–2639 (1981).
Legrand M, Fritig B, Hirth L: Catechol O-methyltransferases of tobacco: evidence for several enzymes with para- and meta-O-methylating activities. FEBS Lett 70: 131–136 (1976).
Lewis NG, Yamamoto E: Lignin: occurrence, biogenesis and biodegradation. Annu Rev Plant Physiol Plant Mol Biol 41: 455–496 (1990).
Montoliu LI, Puigdomènech P, Rigau J: The Tubα3 gene from Zea mays: structure and expression in dividing plant tissues. Gene 94: 201–207 (1990).
Montoliu LI, Rigau J, Puigdomènech P: A tandem of α-tubulin genes preferentially expressed in radicular tissues from Zea mays. Plant Mol Biol 14: 1–15 (1989).
Montoliu LI, Rigau J, Puigdomènech P: Multiple polyadenylation sites are active in the α1-tubulin gene from Zea mays. FEBS Lett 277: 29–32 (1990).
Poulton JE: Transmethylation and demethylation reactions in the metabolism of secondary plant products. In: Stumpf PK, Conn EE (eds) The Biochemistry of Plants: A Comprehensive Treatise, vol. 7: Secondary Plant Products, pp. 667–723. Academic Press, New York (1981).
Poulton JE, Grisebach H, Ebel J, Schaller-Hekeler B, Hahlbrock K: Two distinct S-adenosyl-L-methionine: 3,4-dihydric phenol 3-O-methyltransferases of phenylpropanoid metabolism in soybean cell suspension cultures. Arch Biochem Biophys 173: 301–305 (1976).
Poulton JE, Hahlbrock K, Grisebach H: Enzymatic synthesis of lignin precursors. Arch Biochem Biophys 176: 449–456 (1976).
Queen C, Korn LJ: A comprehensive sequence analysis program for the IBM personal computer. Nucl Acids Res 12: 581–599 (1984).
Rhodes MJC, Hill ACR, Wooltorton LSC: Activity of enzymes involved in lignin biosynthesis in swede root disks. Phytochemistry 15: 707–710 (1976).
Salminen M, Lundström K, Tilgmann C, Savolainen R, Kalkkinen N, Ulmanen I: Molecular cloning and characterization of rat liver catechol-O-methyltransferase. Gene 93: 241–247 (1990).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1989).
Schmitt D, Pakusch A-E, Matern U: Molecular cloning, induction, and taxonomic distribution of caffeoyl-CoA 3-O-methyltransferase, an enzyme involved in disease resistance. J Biol Chem 266: 17416–17423 (1991).
Shimada M, Fushiki H, Higuchi T: O-methyltransferase activity from Japanese black pine. Phytochemistry 11: 2657–2662 (1972).
Shimada M, Kuroda H, Higuchi T: Evidence for the formation of methoxyl groups of ferulic and sinapic acids in Bambusa by the same O-methyltransferase. Phytochemistry 12: 2873–2875 (1973).
Shimada M, Ohashi H, Higuchi T: O-Methyltransferase involved in the biosynthesis of lignins. Phytochemistry 9: 2463–2470 (1970).
Tsang Y-F, Ibrahim RK: Two forms of O-methyltransferase in tobacco cell suspension culture. Phytochemistry 18: 1131–1136 (1979).
Vance CP, Bryan JW: Purification and properties of caffeic acid O-methyltransferase from Alfalfa root nodules. Phytochemistry 20: 41–43 (1981).
Yamada Y, Kuboi T: Significance of caffeic acid-O-methyltransferase in lignification of cultured tobacco cells. Phytochemistry 15: 395–396 (1976).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Collazo, P., Montoliu, L., Puigdomènech, P. et al. Structure and expression of the lignin O-methyltransferase gene from Zea mays L.. Plant Mol Biol 20, 857–867 (1992). https://doi.org/10.1007/BF00027157
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00027157