Abstract
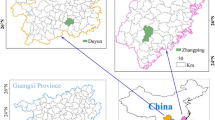
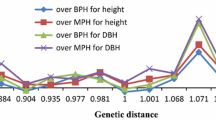
Predicting progeny performance from parental divergence would potentially enhance the efficiency of breeding. Thirteen clones of Masson pine (Pinus massoniana) were crossed in a 4 × 9 tester mating design and 36 full-sib families were generated. There were significant variations in major growth traits and heterosis of growth traits among the 36 full-sib families. A total of 115 alleles were detected with 30 sets of SSR primers. Genetic distances (GDs) among the crossed clonal pairs were calculated based on SSR data. Parental GDs was significantly correlated with all traits evaluated in the full-sib progeny test (P < 0.05), and the correlation coefficient was 0.398, 0.463 and 0.473 for tree height, DBH and wood volume. Parental geographical divergence was also significantly correlated with growth traits of progeny, while the correlation coefficient was lower than that of GDs. These results demonstrated the potential and feasibility of SSR markers for predicting progeny performance of Masson pine. Considering the influence of genotype × environment interaction effect on the stability of full-sib family performance, our results regarding the moderate contribution of parent GDs in predicting the performance of their full-sib progenies is of importance mainly for the specific region where the progeny test was carried out.


Similar content being viewed by others
References
Arcade A, Faivre-rampant P, Le Guerroue B, Pâques LE, Prat D (1996) Heterozygosity and hybrid performance in larch. Theor Appl Genet 93:1274–1281
Beattie AD, Michaels TE, Pauls KP (2003) Predicting progeny performance in common bean (Phaseolus vulgaris L.) using molecular marker-based cluster analysis. Genome 46:259–267
Betrán FJ, Ribaut JM, Beck D, Gonzalez LD (2003) Genetic diversity, specific combining ability, and heterosis in tropical maize under stress and nonstress environments. Crop Sci 43:797–806
Cengel B, Tayanc Y, Kandemir G, Velioglu E, Alan M, Kaya Z (2011) Magnitude and efficiency of genetic diversity captured from seed stands of Pinus nigra (Arnold) subsp. pallasiana in established seed orchards and plantations. New Forest. doi:10.1007/s11056-011-9282-8
Chezhian P, Yasodha R, Ghosh M (2010) Genetic diversity analysis in a seed orchard of Eucalyptus tereticornis. New Forest 40:85–99
Cho Y, Park C, Kwon S (2004) Key DNA markers for predicting heterosis in F1 hybrids of japonica rice. Breed Sci 54:389–397
Dias LA, Picoli EA, Rocha RB, Alfenas AC (2004) A priori choice of hybrid parents in plants. Genet Mol Res 3(3):356–368
Dreisigacker S, Melchinger AE, Zhang P (2005) Hybrid performance and hetrosis in spring bread wheat, and their relations to SSR-based genetic distances and coefficients of parentage. Euphytica 144:51–59
Fofana IJ, Ofori D, Poitel M, Verhaegen D (2009) Diversity and genetic structure of teak (Tectona grandis L.f) in its natural range using DNA microsatellite markers. New Forest 37:175–195
Garcia AAF, Benchimol LL, Barbosa AMM, Geraldi IO (2004) Comparison of RAPD, RFLP, AFLP and SSR markers for diversity studies in tropical maize inbred lines. Genet Mol Biol 27:579–588
Jose MA, Iban E, Silvia A, Pere A (2005) Inheritance mode of fruit traits in melon: heterosis for fruit shape and its correlation with genetic distance. Euphytica 144:31–38
Julfiquar AW, Virmani SS, Carpena AL (1985) Genetic divergence among some maintainer and restorer lines in relation to hybrid breeding in rice (Oryza sativa L.). Theor Appl Genet 70:671–678
Kesawat MS, Kumar BD (2009) Molecular markers: It’s application in crop improvement. J Crop Sci Biotech 12:169–181
Kopp RF, Smart LB, Maynard CA, Tuskan GA, Abrahamson LP (2002) Predicting within-family variability in juvenile height growth of Salix based upon similarity among parental AFLP fingerprints. Theor Appl Genet 105:106–112
Kwon SJ, Ha WG, Wang HG (2002) Relationship between heterosis and genetic divergence in ‘Tongil’-type rice. Plant Breed 121:487–492
Leal AA, Mangolin CA, Amaral AT (2010) Efficiency of RAPD versus SSR markers for determining genetic diversity among popcorn lines. Genet Mol Res 9:9–18
Li Y, Zhang CX (1998) A study on genetic diversity and breeding value of a series of clonal groups in Pinus tabulaef ormis. J Beijing Forestry Univ 20:12–17
Luan F, Sheng Y, Wang Y, Staub JE (2010) Performance of melon hybrids derived from parents of diverse geographic origins. Euphytica 173:1–16
Ma KH, Kim NS, Lee GA, Lee SY, Lee JK (2009) Development of SSR markers for studies of diversity in the genus Fagopyrum. Theor Appl Genet 119:1247–1254
Mohapatra KP, Sehgal RN, Sharma RK, Mohapatra T (2009) Genetic analysis and conservation of endangered medicinal tree species Taxus wallichiana in the Himalayan region. New Forest 37:109–121
Moriguchi Y, Yamazaki Y, Taira H, Tsumura Y (2010) Mating patterns in an indoor miniature Cryptomeria japonica seed orchard as revealed by microsatellite markers. New Forest 39:261–273
Muro-Abad JI, Rocha RB, Cruz CD, Araújo EF (2005) Obtainment of Eucalyptus spp. hybrids aided by molecular markers-SSR analysis. Scientia Forestalis 67:53–63
Naik D, Singh D, Vartak V, Paranjpe S, Bhargava S (2009) Assessment of morphological and genetic diversity in Gmelina arborea Roxb. New Forest 38:99–115
Nkongolo KK (1999) RAPD and cytological analyses of Picea spp. from different provenances: Genomic relationships among taxa. Hereditas 130:137–144
Qin GF (2002) Geographical origin and evolution of Masson pine. Forest Res (In Chinese) 15:406–412
Reif JC, Melchinger AE, Xia XC (2003) Use of SSRs for establishing heterotic groups in subtropical maize. Theor Appl Genet 107:947–957
Riday H, Brummer E (2006) Dissection of heterosis in alfalfa hybrids. Dev Plant Breed 11:315–324
Shieh GJ, Thseng FS (2002) Genetic diversity of Tainan-white maize inbred lines and prediction of single cross hybrid performance using RAPD markers. Euphytica 124:307–313
Teklewold A, Becker HC (2006) Comparison of phenotypic and molecular distances to predict heterosis and F1 performance in Ethiopian mustard (Brassica carinata A. Braun). Theor Appl Genet 112:752–759
Venkateswarlu M, Raje-Urs S, Surendra-Nath B (2006) A first genetic linkage map of mulberry (Morus spp.) using RAPD, ISSR, and SSR markers and pseudo test cross mapping strategy. Tree Genet Genomes 3:15–24
Vijayan K, Chatterjee SN (2003) ISSR profiling of Indian cultivars of mulberry (Morus spp.) and its relevance to breeding programs. Euphytica 131:53–63
Wee AKS, Li C, Dvorak WS, Hong Y (2011) Genetic diversity in natural populations of Gmelina arborea: implications for breeding and conservation. New Forest. doi:10.1007/s11056-011-9288-2
Weston FM, Akinnifesi FK, Stedje B (2010) Genetic diversity within and among southern African provenances of Uapaca kirkiana Muell. Arg using morphological and AFLP markers. New Forest 40:383–399
Yu CY, Hu SW, Zhao HX, Guo AG (2005) Genetic distances revealed by morphological characters, isozymes, protein and RAPD markers and their relationships with Hybrid performance in oilseed rape (Brassica napus L.). Theor Appl Genet 110:511–518
Acknowledgments
We thank the editors and the reviewers for their hard work in the improvement of this paper. We thank the members of the Forest Breeding Group of the Research Institute of Subtropical Forestry for discussion, encouragement, and assistance in the traits measurements. We thank the staff of the Laoshan forest farm of Chunan county, Zhejiang Province for their work in field trials. This research was supported by a research project from the National High Technology Research and Development Program of China (“863” Program) (2011AA100203), the National Key Technology R&D Program in the 12th Five year Plan of China (2012BAD01B02), the National Natural Science Foundation financed by the Chinese Government (31070599) and the National Department Public Benefit Research Foundation (No. 201104010) from Chinese government.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, Y., Yang, Q., Zhou, Z. et al. Divergence among Masson pine parents revealed by geographical origins and SSR markers and their relationships with progeny performance. New Forests 44, 341–355 (2013). https://doi.org/10.1007/s11056-012-9340-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11056-012-9340-x