Abstract
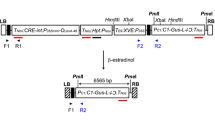
Curcin, a type I ribosomal inhibiting protein-RIP, encoded by curcin precursor gene, is a phytotoxin present in Jatropha (Jatropha curcas L.). Here, we report designing of RNAi construct for the curcin precursor gene and further its genetic transformation of Jatropha to reduce its transcript expression. Curcin precursor gene was first cloned from Jatropha strain DARL-2 and part of the gene sequence was cloned in sense and antisense orientation separated by an intron sequence in plant expression binary vector pRI101 AN. The construction of the RNAi vector was confirmed by double digestion and nucleotide sequencing. The vector was then mobilized into Agrobacterium tumefaciens strain GV 3101 and used for tissue culture independent in planta transformation protocol optimized for Jatropha. Germinating seeds were injured with a needle before infection with Agrobacterium and then transferred to sterilized sand medium. The seedlings were grown for 90 days and genomic DNA was isolated from leaves for transgenic confirmation based on real time PCR with NPT II specific dual labeled probe. Result of the transgenic confirmation analysis revealed presence of the gene silencing construct in ten out of 30 tested seedlings. Further, quantitative transcript expression analysis of the curcin precursor gene revealed reduction in the transcript abundance by more than 98 % to undetectable level. The transgenic plants are being grown in containment for further studies on reduction in curcin protein content in Jatropha seeds.





Similar content being viewed by others
References
Openshaw K (2000) A review of Jatropha curcas: an oil plant unfulfilled promise. Biomass Bioenergy 19:1–15
Nambisan P (2007) Biotechnological interventions in Jatropha for biodiesel production. Curr Sci 93:1347–1348
Kywe TT, Oo MM (2009) Production of biodiesel from Jatropha oil (Jatropha curcas) in pilot plant. World Academy Sci Eng Technol 50:477–483
King AJ, He W, Cuevas JA, Freudenberger M, Ramiaramanana D, Graham IA (2009) Potential of Jatropha curcas as a source of renewable oil and animal feed. J Exp Bot 60:2897–2905
Barbieri L, BattelliM Stirpe F (1993) Ribosome-inactivating protein from plants. Biochim Biophy Acta 1154:237–282
Lin J, Chen Y, Xu Y, Yan F, Tang L, Chen F (2003) Cloning and expression of curcin, a ribosome-inactivating protein from the seeds of Jatropha curcas. Acta Bot Sin 45:858–863
Qin W, Huang M-X, Ying X, Zhang X-S, Fang C (2005) Expression of a ribosome inactivating protein (curcin 2) in Jatropha curcas is induced by stress. J Biosci 30:351–357
Martı′nez-Herrera J, Siddhuraju P, Francis G, Davila-Ortiz G, Becker K (2006) Chemical composition, toxic/antimetabolic constituents, and effects of different treatments on their levels, in four provenances of Jatropha curcas L. from Mexico. Food Chem 96:80–89
He W, King AJ, Khan MA, Cuevas JA, Ramiaramanana D, Graham IA (2011) Analysis of seed phorbol-ester and curcin content together with genetic diversity in multiple provenances of Jatropha curcas L. from Madagascar and Mexico. Plant Physiol Biochem 49:1183–1190
Baulcombe D (2004) RNA silencing in plants. Nature 431:356–363
Ye J, Qu J, Bui HTN, Chua NH (2009) Rapid analysis of Jatropha curcas gene functions by virus-induced gene silencing. Plant Biotechnol J 7:964–976
Zhong CY, Li FW, Hui ZM, Cheng XQ. 2010. Jatropha curcas curcin genes, tissue-specific promoters and generation of curcin-deficient transgenic Jatropha plants. WIPO Patent Application SG2010/000206, 2010
Travella S, Ross SM, Harden J, Everett C, Snape JW, Harwood WA (2005) A comparison of transgenic barley lines produced by particle bombardment and Agrobacterium mediated techniques. Plant Cell Rep 23:780–789
Deore AC, Johnson ST (2008) High-frequency plant regeneration from leaf-disc cultures of Jatropha curcas L.: an important biodiesel plant. Plant Biotechnol Rep 2:7–11
Kumar N, Vijay AKG, Sudheer PDVN, Sarkar T, Reddy MP, Radhakrishnan T, Kaul T, Reddy MK, Sopori SK (2010) Stable genetic transformation of Jatropha curcas via Agrobacterium tumefaciens-mediated gene transfer using leaf explants. Ind Crops Prod 32:41–47
Sujatha M, Makkar HPS, Becker K (2005) Shoot bud proliferation from axillary nodes and leaf sections of non-toxic Jatropha curcas L. Plant Growth Regul 47:83–90
Datta MM, Mukherjee P, Ghosh B, Jha TB (2007) In vitro clonal propagation of biodiesel plant (Jatropha curcas L.). Curr Sci 93:1438–1442
Thepsamran N, Thepsithar C, Thongpukdee A (2008) In vitro induction of shoots and roots from Jatropha curcas L. explants. J Horticult Sci Biotechnol 83:106–112
Li M, Li H, Jiang H, Pan X, Wu G (2008) Establishment of an Agrobacteriuim-mediated cotyledon disc transformation method for Jatropha curcas. Plant Cell Tissue Org Cult 92:173–181
Keshamma E, Sreevathsa R, Manoj Kumar A, Reddy KN, Manjulatha M, Shanmugam NB, Kumar ARV, Udayakumar M (2012) Agrobacterium-mediated in planta transformation of field bean (Lablab purpureus L.) and recovery of stable transgenic plants expressing the cry1AcF gene. Plant Mol Biol Rep 30:67–78
Bent AF (2000) Arabidopsis in planta transformation. Uses, mechanisms, and prospects for transformation of other species. Plant Physiol 124:1540–1547
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Pagni M, Ioannidis V, Cerutti L, Zahn-Zabal M, Jongeneel CV, Hau J, Martin O, Kuznetsov D, Falquet L (2007) MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res 35:W433–W437
Patade VY, Bhargava S, Suprasanna P (2012) Transcript expression profiling of stress responsive genes in response to short-term salt or PEG stress in sugarcane leaves. Mol Biol Rep 39:3311–3318
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologists programmers. In: Krawetzs S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using a real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL (2011) “ViennaRNA package 2.0″. Algorithms Mol Biol. doi:10.1186/1748-7188-6-26
Makkar HPS, Becker K (1999) Nutritional studies on rats and fish (carp Cyprinus carpio) fed diets containing unheated and heated Jatropha curcas meal of a non-toxic provenanace. Plant Foods Hum Nutr 53:183–192
Rakshit KD, Darukeshwara J, Raj KRR, Narasimhamurthy K, Saibaba P, Bhagya S (2008) Toxicity studies of detoxified Jatropha meal (Jatropha curcas) in rats. Food Chem Toxicol 46:3621–3625
Yin, ZC, Wu, Li Fang, Mao HZ, Qiu CX (2010) Jatropha curcas curcin genes, tissue-specific promoters and generation of curcin-deficient transgenic Jatropha plants. WIPO Patent Application WO/2010/140981, 2010
Huang M-X, Hou P, Wei Q, Xu Y, Chen F (2008) A ribosome-inactivating protein (curcin 2) induced from Jatropha curcas can reduce viral and fungal infection in transgenic tobacco. Plant Growth Regul 54:115–123
Palle SR, Campbell LM, Pandeya D, Puckhaber L, Tollack LK, Marcel S, Sundaram S, Stipanovic RD, Wedegaertner TC, Hinze L, Rathore KS (2013) RNAi-Mediated ultra-low gossypol cottonseed trait: performance of transgenic lines under field conditions. Plant Biotechnol J 11:296–304
Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, Robinson SP, Gleave AP, Green AG, Waterhouse PM (2001) Construct design for efficient, effective and highthroughput gene silencing in plants. Plant J 27:581–590
Acknowledgments
Research fellowship from DRDO to Deepti Khatri and Kamal Kumar is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Patade, V.Y., Khatri, D., Kumar, K. et al. RNAi Mediated curcin precursor gene silencing in Jatropha (Jatropha curcas L.). Mol Biol Rep 41, 4305–4312 (2014). https://doi.org/10.1007/s11033-014-3301-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3301-8