Abstract
Studies have demonstrated that changes in DNA methylation of cancer related genes can be an elementary process accounting for ovarian tumorigenesis. Therefore, we evaluated the possible association of single nucleotide polymorphisms (SNPs) of DNA methyltransferases (DNMTs) genes, including DNMT1, DNMT3B, and DNMT3A, with ovarian cancer development in the Polish population. Using PCR–RFLP and HRM analyses, we studied the prevalence of the DNMT1 rs8101626, rs2228611 and rs759920, DNMT3A rs2289195, 7590760, rs13401241, rs749131 and rs1550117, and DNMT3B rs1569686, rs2424913 and rs2424932 SNPs in patients with ovarian cancer (n = 159) and controls (n = 180). The lowest p values of the trend test were observed for the DNMT1 rs2228611 and rs759920 SNPs in patients with ovarian cancer (p trend = 0.0118 and p trend = 0.0173, respectively). Moreover, we observed, in the recessive inheritance model, that the DNMT1 rs2228611 and rs759920 SNPs are associated with an increased risk of ovarian cancer development [OR 1.836 (1.143–2.949), p = 0.0114, p corr = 0.0342, and OR 1.932 (1.185–3.152), p = 0.0078, p cor=0.0234, respectively]. However, none of other nine studied SNPs displayed significant contribution to the development of ovarian cancer. Furthermore, haplotype and multifactor dimensionality reduction analysis of the studied DNMT1, DNMT3B, and DNMT3A polymorphisms did not reveal either SNP combinations or gene interactions to be associated with the risk of ovarian cancer development. Our results may suggest that DNMT1 variants may be risk factors of ovarian cancer.
Similar content being viewed by others
Introduction
Ovarian cancer includes any malignant growth that may develop in disparate parts of the ovary; however, the majority of ovarian malignancies arise from the ovarian epithelium [1]. This type of cancer usually exhibits vague symptoms and is the one of the most lethal gynecological cancers in women in the United States and Europe [2]. Although ovarian cancer has been intensively studied, the underlying cause of this malignant disease remains undetermined [1, 3]. There is a lot of evidence that shows a reduced risk for ovarian cancer in women who use oral contraceptives, have greater parity, or breastfed long-term [3]. Known risk factors of ovarian cancer include infertility, early age of menarche, late age of menopause, inflammation, some environmental factors, and genetic background [3]. The known genetic factors encompass a few high-penetrance genes (e.g., BRCA1) and numerous moderate and low-penetrance ovarian cancer susceptibility genes [3]. However, these factors alone are not sufficient for ovarian tumorigenesis, which indicates that attention must be paid to the role of epigenetic status changes during ovarian carcinogenesis [4, 5].
The term epigenetics describes the reversible regulation of gene expression that does not occur due to changes in the DNA sequence [6, 7]. Epigenetic traits include DNA methylation, covalent modification of histones, and expression status of genes modulated by μRNA [6, 7].
DNA methylation in mammalian cells is typically restricted to covalent modification, in which a methyl group is added to a cytosine located in CpG dinucleotides in the genomic DNA [6]. CpG dinucleotides are frequently situated in rich CpG genomic sites designated as CpG islands, which are found in approximately half of all genes in humans [8]. Hypomethylation of regulatory DNA sequences increases the expression of proto-oncogenes and genes encoding proteins involved in genomic instability and malignant cell growth and metastasis [6, 9]. By contrast, hypermethylation of the promoter regions of tumor suppressor genes (TSGs) causes the transcriptional silencing of TSGs [6, 9].
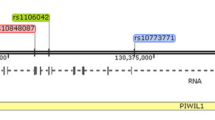
There are three types of enzymatically active DNA methyltransferases (DNMTs), designated DNMT1, DNMT3A, and DNMT3B [6, 9]. Abnormal levels of DNMT1, DNMT3A and DNMT3B contributing to changes in the expression of cancer related genes have been found in different types of malignancies [10–13]. The DNMTs levels can be changed by single nuclear polymorphisms (SNPs) situated within their genes and which may affect the development of various cancers [14, 15]. Therefore, we selected 11 SNPs of DNMT1, DNMT3A and DNMT3B located in distinct blocks of linkage disequilibrium (LD) according to HapMap CEU data (http://hapmap.ncbi.nlm.nih.gov/) (Supplemental Table 1; Supplemental Fig. 1a–c). Furthermore, we aimed to study whether these DNMT1, DNMT3A and DNMT3B SNPs can be a genetic risk factor of ovarian cancer.
Materials and methods
Patients and controls
The patients include 159 women with histologically recognized ovarian carcinoma according to the International Federation of Gynecology and Obstetrics (FIGO). Histopathological classification, including the stage, grade and tumor type, was performed by an experienced pathologist (Table 1). The controls included 180 unrelated healthy female volunteers who were matched by age to the cancer patients (Table 1). Written informed consent was obtained from all participating individuals. The procedures of the study were approved by the Local Ethical Committee of Poznań University of Medical Sciences. All women with ovarian cancer and controls were Caucasian from the Wielkopolska area of Poland.
Genotyping
Genomic DNA was isolated from peripheral blood leucocytes by salt extraction. DNA samples were genotyped for the 11 SNPs in DNMT1, DNMT3A and DNMT3B (Supplemental Table 1; Supplemental Fig. 1a–c.). SNPs were selected with the use of the genome browsers of the International HapMap Consortium (http://www.hapmap.org/index.html.en), UCSC (http://genome.ucsc.edu) and dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/). SNPs were selected based on functional significance, location in different LD blocks, and minor allele frequency (MAF) >0.1 in the Caucasian population. Genotyping of the DNMT1 rs2228611, rs759920, DNMT3A rs2289195, rs13401241, rs749131, rs1550117, DNMT3B rs2424932 SNPs was performed by high resolution melting curve analysis (HRM) on the LightCycler 480 system (Roche Diagnostics, Mannheim, Germany). Genotyping of the DNMT1 rs8101626, DNMT3A rs7590760, and DNMT3B rs1569686, rs2424913SNPs was conducted by PCR, followed by the appropriate restriction enzyme digestion (PCR–RFLP) according to the manufacturer’s instructions (Fermentas, Vilnius, Lithuania). DNA fragments were separated in 2 % agarose gels and visualized by ethidium bromide staining. Primer sequences and conditions for HRM and PCR–RFLP analyses are presented in Supplemental Table 2. Genotyping quality was evaluated by repeated genotyping of 10 % randomly selected samples.
Statistical analysis
For each SNP, the Hardy–Weinberg equilibrium (HWE) was assessed by Pearson’s goodness-of-fit Chi square (χ 2) statistic. The differences in the allele and genotype frequencies between cases and controls were determined using standard χ 2 or Fisher tests. SNPs were tested for association with ovarian cancer using the Cochran-Armitage trend test. The odds ratio (OR) and associated 95 % confidence intervals (95 %CI) were also calculated. The data were analyzed under recessive and dominant inheritance models. To adjust for the multiple testing, we employed a correction factor of 3 (0.005/3 = 0.0167) to take into consideration the number of genes evaluated. This represents a generally accepted correction strategy; one not as stringent as the Bonferroni correction, yet still taking the number of totally independent analyses into account. Pair-wise LD between selected SNPs was computed as both D′ and r 2 values using HaploView 4.0 software (Broad Institute, Cambridge, MA). Haplotype analysis was performed using the UNPHASED 3.1.5 program with the following analysis options: all window sizes, full model and uncertain haplotype option [16]. Haplotypes with a frequency below 0.01 were set to zero. The p values for both global and individual tests of haplotype distribution between cases and controls were determined. Statistical significance was assessed using the 1,000-fold permutation test.
High order gene–gene interactions among all tested polymorphic loci were studied by the multifactor dimensionality reduction (MDR) approach (MDR version 2.0 beta 5). A detailed explanation on the MDR method has been described elsewhere [17]. Based on the obtained testing balanced accuracy and cross-validation consistency values, the best statistical gene–gene interaction models were established. A 1,000-fold permutation test was used to assess the statistical significance of MDR models (MDR permutation testing module 0.4.9 alpha).
Results
Association of DNMT1, DNMT3A and DNMT3B SNPs with ovarian cancer development
The frequency of all studied genotypes did not exhibit deviation from the HWE between all investigated groups (p > 0.05). The lowest (0.08) and highest (0.49) MAF found for the control samples was for DNMT3A rs1550117 and DNMT1 rs2228611 SNPs, respectively (Table 2). The number of genotypes, OR, and 95 % CI calculations for the 11 studied DNMT1, DNMT3B, and DNMT3A SNPs are presented in Table 2. The lowest p values of the trend test were observed for the DNMT1 rs2228611 and rs759920 SNPs in patients with ovarian cancer (p trend = 0.0118 and p trend = 0.0173, respectively) (Table 2). The statistical significance for multiple testing determined by correction of genes number was p = 0.0167. Therefore, we observed, in a recessive inheritance model, that DNMT1 rs2228611 and rs759920 SNPs are associated with increased risk of ovarian cancer development [OR 1.836 (1.143–2.949), p = 0.0114, and OR 1.932 (1.185–3.152), p = 0.0078, respectively] (Table 2). However, none of the other nine studied DNMT1, DNMT3B and DNMT3A SNPs exhibited significant association either in dominant or recessive inheritance models with ovarian cancer development (Table 2).
Association of DNMT1, DNMT3B, and DNMT3A haplotypes with ovarian cancer development
Haplotype analysis of DNMT1, DNMT3B, and DNMT3A polymorphisms did not reveal SNP combinations associated with the risk of ovarian cancer development (Table 3). In patients, the lowest global p = 0.0502 was observed for haplotypes composed of the DNMT1 rs2228611 and rs759920 SNPs (Table 3). However, these results were not statistically significant when permutations were used to generate empiric p values. The empirical 5 % quintile of the best p value after 1,000 permutations was 0.002566. The selected 11 SNPs situated in distinct regions of DNMT1, DNMT3B, and DNMT3A were either in weak or strong pairwise LD. This was calculated from the control samples, and had D′ ranges of 0.934–0.976 for DNMT1 SNPs, 0.109–1.000 for DNMT3A SNPs, and 0.626–0.943 for DNMT3B SNPs (Supplementary Table 3).
MDR analysis of gene–gene interactions among the studied DNMT1, DNMT3A and DNMT3B SNPs
Exhaustive MDR analysis evaluating two- to four-loci combinations of all studied SNPs for each comparison did not reveal statistical significance in predicting susceptibility to ovarian cancer development (Table 4). The best combination of possibly interactive polymorphisms was observed for rs759920 of DNMT1, rs2289195 and rs7590760 of DNMT3A and rs2424932 of DNMT3B (testing balanced accuracy = 59.03 %, cross validation consistency of eight out of ten, permutation test p = 0.0940).
Discussion
As with other cancer types, aberrant DNA methylation encompassing CpG island hypermethylation and global hypomethylation of heterochromatin has been demonstrated in ovarian cancer [4, 5, 12, 18]. To date, it several genes have been found to be down-regulated in ovarian cancer due to their hypermethylation [19–23]. These genes include classical TSGs and genes encoding cell adhesion molecules, proapoptotic proteins, and other proteins supporting ovarian tumorigenesis [19–23]. DNA hypomethylation in ovarian cancer has been found in chromosome 1 satellite 2 and long interspersed nuclear element-1 (LINE-1) repetitive elements [18, 24]. In addition to these findings, many genes are hypomethylated and overexpressed in this type of cancer. Among these are genes encoding activators of the mitogen-activated protein kinases, insulin-like growth factor-2, and proteins associated with chemoresistance [18, 25–27].
Increased levels of some DNMTs have been observed in ovarian cancer cell lines and primary ovarian cancerous tissues as compared to normal ovarian cells [10, 11, 28]. Ahluwalia et al. [10] found significantly higher DNMT1 and DNMT3B mRNA levels in ovarian cancer cells than in normal ovarian surface epithelial cells. Moreover, Chen et al. [28] observed significantly elevated DNMT1 and DNMT3B mRNA levels in primary and recurrent epithelial ovarian carcinoma compared to normal epithelial ovarian cells. Recently, Cheng et al. (2011) demonstrated that DNMT1, but not DNMT3A or DNMT3B, levels were increased in serous borderline ovarian tumor cells. They also demonstrated that DNMT1 levels correlated with E-cadherin promoter methylation [11].
In our study we found that the DNMT1 rs2228611 and rs759920 SNPs can be risk factors of ovarian cancer in a Polish population. To date, the DNMT1 rs2228611 SNP has been associated with sporadic infiltrating ductal breast carcinoma among Chinese Han women in the Heilongjiang Province [29]. Moreover, the DNMT1 rs2228611 SNP modified associations between urinary cadmium and hypomethylation of LINE-1 repeated sequences [30]. In addition to these findings, the two intronic polymorphisms rs2241531 and rs4804490 of DNMT1 may be associated with hepatitis B virus (HBV) clearance and protection from the development of hepatocellular carcinoma [31].
Our studies did not demonstrate an association of the DNMT3B rs1569686, rs2424913 and rs2424932 SNPs with ovarian cancer. To date, the DNMT3B rs2424913 polymorphism has been demonstrated to be a risk factor of lung, breast, and head and neck cancers and age of onset of hereditary nonpolyposis colorectal cancer (CRC) [32–35]. Furthermore, the DNMT3B rs2424913 SNP may contribute to reduced methylation of CpG islands of the MYOD and MLH1 genes in normal colonic mucosa of patients with CRC, and with increased promoter methylation of TSGs related to the development of prostate cancer [36, 37]. Another polymorphism of DNMT3B, rs2424909, being in great LD with rs2424913 has been found to be a genetic risk factor of gastric cancer, CRCs, and gallbladder carcinoma [38–40].
It has recently been found that the DNMT3A polymorphism rs1550117 alters the promoter activity and risk of gastric cancer and CRC [14, 15]. However, we did not find an association of any of the five studied DNMT3A SNPs with ovarian cancer development. Frequent mutations in DNMT3A have been demonstrated in human T cell lymphoma, myelodysplastic syndromes, as well as myeloid and monocytic leukemias [41–44]. Moreover, the DNMT3A intronic polymorphism rs13428812, situated in the same LD block with our studied rs7590760, has been shown to contribute to Crohn’s disease [45].
DNMT3A and DNMT3B carry out de novo methylation [7, 13]. DNMT3A and DNMT3B are involved in imprinting of germ cells and normal embryonic development [46, 47]. Moreover, DNMT3A contributes to the differentiation of neural progenitors and hematopoietic stem cells [48, 49]. DNMT1 is responsible for the maintenance of the hemimethylated DNA methylation pattern during DNA replication, and also has a role in gene silencing [13, 50].
Our study may suggest that Polish women bearing either the DNMT1 rs2228611 or rs759920 SNP may have an increased risk of ovarian cancer. Neither of these polymorphisms is a functional variant, and their association with ovarian cancer may be due to LD with one or more functional polymorphisms of DNMT1. Therefore, to confirm the role of these SNPs in ovarian cancer, this study should be repeated in a larger and independent cohort, and functional studies of these SNPs must be conducted. Moreover, a future genome-wide SNPs genotyping study in our sample collection will additionally provide more knowledge of the genes contributing to the onset of this disease.
References
Kuhn E, Kurman RJ, Shih IM (2012) Ovarian cancer is an imported disease: fact or fiction? Curr Obstet Gynecol Rep 1:1–9
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ (2008) Cancer statistics, 2008. CA Cancer J Clin 58:71–96
Sueblinvong T, Carney ME (2009) Current understanding of risk factors for ovarian cancer. Curr Treat Options Oncol 10:67–81
Barton CA, Hacker NF, Clark SJ, O’Brien PM (2008) DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol Oncol 109:129–139
Balch C, Fang F, Matei DE, Huang TH, Nephew KP (2009) Minireview: epigenetic changes in ovarian cancer. Endocrinology 150:4003–4011
Goldberg AD, Allis CD, Bernstein E (2007) Epigenetics: a landscape takes shape. Cell 128:635–638
Turek-Plewa J, Jagodziński PP (2005) The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett 10:631–647
Antequera F, Bird A (1993) Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA 90:11995–11999
Li Y, Tollefsbol TO (2010) Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr Med Chem 17:2141–2151
Ahluwalia A, Hurteau JA, Bigsby RM, Nephew KP (2001) DNA methylation in ovarian cancer. II. Expression of DNA methyltransferases in ovarian cancer cell lines and normal ovarian epithelial cells. Gynecol Oncol 82:299–304
Cheng JC, Auersperg N, Leung PC (2011) Inhibition of p53 represses E-cadherin expression by increasing DNA methyltransferase-1 and promoter methylation in serous borderline ovarian tumor cells. Oncogene 30:3930–3942
Taberlay PC, Jones PA (2011) DNA methylation and cancer. Prog Drug Res 67:1–23
Sowińska A, Jagodzinski PP (2007) RNA interference-mediated knockdown of DNMT1 and DNMT3B induces CXCL12 expression in MCF-7 breast cancer and AsPC1 pancreatic carcinoma cell lines. Cancer Lett 255:153–159
Fan H, Liu D, Qiu X, Qiao F, Wu Q, Su X, Zhang F, Song Y, Zhao Z, Xie W (2010) A functional polymorphism in the DNA methyltransferase-3A promoter modifies the susceptibility in gastric cancer but not in esophageal carcinoma. BMC Med 8:12
Zhao Z, Li C, Song Y, Wu Q, Qiao F, Fan H (2012) Association of the DNMT3A -448A>G polymorphism with genetic susceptibility to colorectal cancer. Oncol Lett 3:450–454
Dudbridge F (2008) Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered 66:87–98
Hahn LW, Ritchie MD, Moore JH (2003) Multifactor dimensionality reduction software for detecting gene–gene and gene-environment interactions. Bioinformatics 19:376–382
Widschwendter M, Jiang G, Woods C, Müller HM, Fiegl H, Goebel G, Marth C, Müller-Holzner E, Zeimet AG, Laird PW, Ehrlich M (2004) DNA hypomethylation and ovarian cancer biology. Cancer Res 64:4472–4480
Press JZ, De Luca A, Boyd N, Young S, Troussard A, Ridge Y, Kaurah P, Kalloger SE, Blood KA, Smith M, Spellman PT, Wang Y, Miller DM, Horsman D, Faham M, Gilks CB, Gray J, Huntsman DG (2008) Ovarian carcinomas with genetic and epigenetic BRCA1 loss have distinct molecular abnormalities. BMC Cancer 8:17
Milde-Langosch K, Ocon E, Becker G, Löning T (1998) p16/MTS1 inactivation in ovarian carcinomas: high frequency of reduced protein expression associated with hyper-methylation or mutation in endometrioid and mucinous tumors. Int J Cancer 79:61–65
Pruitt K, Ulkü AS, Frantz K, Rojas RJ, Muniz-Medina VM, Rangnekar VM, Der CJ, Shields JM (2005) Ras-mediated loss of the pro-apoptotic response protein Par-4 is mediated by DNA hypermethylation through Raf-independent and Raf-dependent signaling cascades in epithelial cells. J Biol Chem 280:23363–23370
Feng W, Marquez RT, Lu Z, Liu J, Lu KH, Issa JP, Fishman DM, Yu Y, Bast RC Jr (2008) Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer 112:1489–1502
Arnold JM, Cummings M, Purdie D, Chenevix-Trench G (2001) Reduced expression of intercellular adhesion molecule-1 in ovarian adenocarcinomas. Br J Cancer 85:1351–1358
Pattamadilok J, Huapai N, Rattanatanyong P, Vasurattana A, Triratanachat S, Tresukosol D, Mutirangura A (2008) LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int J Gynecol Cancer 18:711–717
Strathdee G, Vass JK, Oien KA, Siddiqui N, Curto-Garcia J, Brown R (2005) Demethylation of the MCJ gene in stage III/IV epithelial ovarian cancer and response to chemotherapy. Gynecol Oncol 97:898–903
Murphy SK, Huang Z, Wen Y, Spillman MA, Whitaker RS, Simel LR, Nichols TD, Marks JR, Berchuck A (2006) Frequent IGF2/H19 domain epigenetic alterations and elevated IGF2 expression in epithelial ovarian cancer. Mol Cancer Res 4:283–292
Balch C, Huang TH, Brown R, Nephew KP (2004) The epigenetics of ovarian cancer drug resistance and resensitization. Am J Obstet Gynecol 191:1552–1572
Chen CL, Yan X, Gao YN, Liao QP (2005) Expression of DNA methyltransferase 1, 3A and 3B mRNA in the epithelial ovarian carcinoma. Zhonghua Fu Chan Ke Za Zhi 40:770–774
Xiang G, Zhenkun F, Shuang C, Jie Z, Hua Z, Wei J, Da P, Dianjun L (2010) Association of DNMT1 gene polymorphisms in exons with sporadic infiltrating ductal breast carcinoma among Chinese Han women in the Heilongjiang Province. Clin Breast Cancer 10:373–377
Hossain MB, Vahter M, Concha G, Broberg K (2012) Low-level environmental cadmium exposure is associated with DNA hypomethylation in Argentinean women. Environ Health Perspect 120:879–884
Chun JY, Bae JS, Park TJ, Kim JY, Park BL, Cheong HS, Lee HS, Kim YJ, Shin HD (2009) Putative association of DNA methyltransferase 1 (DNMT1) polymorphisms with clearance of HBV infection. BMB Rep 42:834–839
Liu Z, Wang L, Wang LE, Sturgis EM, Wei Q (2008) Polymorphisms of the DNMT3B gene and risk of squamous cell carcinoma of the head and neck: a case–control study. Cancer Lett 268:158–165
Shen H, Wang L, Spitz MR, Hong WK, Mao L, Wei Q (2002) A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer Res 62:4992–4995
Montgomery KG, Liu MC, Eccles DM, Campbell IG (2004) The DNMT3B C–>T promoter polymorphism and risk of breast cancer in a British population: a case–control study. Breast Cancer Res 6:R390–R394
Jones JS, Amos CI, Pande M, Gu X, Chen J, Campos IM, Wei Q, Rodriguez-Bigas M, Lynch PM, Frazier ML (2006) DNMT3b polymorphism and hereditary nonpolyposis colorectal cancer age of onset. Cancer Epidemiol Biomarkers Prev 15:886–891
Singal R, Das PM, Manoharan M, Reis IM, Schlesselman JJ (2005) Polymorphisms in the DNA methyltransferase 3b gene and prostate cancer risk. Oncol Rep 14:569–573
Kawakami K, Ruszkiewicz A, Bennett G, Moore J, Grieu F, Watanabe G, Iacopetta B (2006) DNA hypermethylation in the normal colonic mucosa of patients with colorectal cancer. Br J Cancer 94:593–598
Hu J, Fan H, Liu D, Zhang S, Zhang F, Xu H (2010) DNMT3B promoter polymorphism and risk of gastric cancer. Dig Dis Sci 55:1011–1016
Srivastava K, Srivastava A, Mittal B (2010) DNMT3B -579 G>T promoter polymorphism and risk of gallbladder carcinoma in North Indian population. J Gastrointest Cancer 41:248–253
Bao Q, He B, Pan Y, Tang Z, Zhang Y, Qu L, Xu Y, Zhu C, Tian F, Wang S (2011) Genetic variation in the promoter of DNMT3B is associated with the risk of colorectal cancer. Int J Colorectal Dis 26:1107–1112
Couronné L, Bastard C, Bernard OA (2012) TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med 366:95–96
Kolquist KA, Schultz RA, Furrow A, Brown TC, Han JY, Campbell LJ, Wall M, Slovak ML, Shaffer LG, Ballif BC (2011) Microarray-based comparative genomic hybridization of cancer targets reveals novel, recurrent genetic aberrations in the myelodysplastic syndromes. Cancer Genet 204:603–628
Marková J, Michková P, Burčková K, Březinová J, Michalová K, Dohnalová A, Maaloufová JS, Soukup P, Vítek A, Cetkovský P, Schwarz J (2012) Prognostic impact of DNMT3A mutations in patients with intermediate cytogenetic risk profile acute myeloid leukemia. Eur J Haematol 88:128–135
Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, Liang WX, Mi JQ, Song HD, Li KQ, Chen Z, Chen SJ (2011) Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet 43:309–315
Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wijmenga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Büning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, De Vos M, D’Inca R, Dubinsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van Gossum A, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Panés J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D’Amato M, Weersma RK, Kugathasan S, Griffiths AM, Mansfield JC, Vermeire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M (2010) Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet 42:1118–1125
Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H (2004) Essential role for de novo DNA methyltransferase DNMT3A in paternal and maternal imprinting. Nature 429:900–903
Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases DNMT3A and DNMT3B are essential for de novo methylation and mammalian development. Cell 99:247–257
Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, Liang S, Lu Y, Darlington GJ, Meissner A, Issa JP, Godley LA, Li W, Goodell MA (2011) DNMT3A is essential for hematopoietic stem cell differentiation. Nat Genet 44:23–31
Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE (2010) Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329:444–448
Dhe-Paganon S, Syeda F, Park L (2011) DNA methyl transferase 1: regulatory mechanisms and implications in health and disease. Int J Biochem Mol Biol 2:58–66
Acknowledgments
Supported by Grant No. 502-01-01124182-07474, Poznan University of Medical Sciences.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. 1
The linkage disequilibrium (LD) plot of HapMap SNPs within the DNMT1 (a), DNMT3A (b) and DNMT3B (c) regions. The plot was generated using the genotype data from HapMap CEU samples and the Haploview 4.0 software (Broad Institute, Cambridge, MA). The names of the examined SNPs are enclosed in boxes. The numbers in the squares indicate percentage of LD between a given pair of SNPs (D′ values). (DOC 21 kb)
Table 1
Characteristics of polymorphisms genotyped in the data set (DOC 35 kb)
Table 2
HRM and RFLP conditions for the identification of polymorphisms genotyped in the data set (DOC 56 kb)
Table 3
Linkage disequilibrium between markers of the DNMT1 gene in the control samples. Linkage disequilibrium between markers of the DNMT3A gene in the control samples (DOC 37 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Mostowska, A., Sajdak, S., Pawlik, P. et al. DNMT1, DNMT3A and DNMT3B gene variants in relation to ovarian cancer risk in the Polish population. Mol Biol Rep 40, 4893–4899 (2013). https://doi.org/10.1007/s11033-013-2589-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-013-2589-0