Abstract
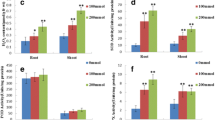
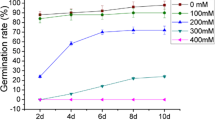
A proteomics approach was used to analyze the response mechanism in soybean seedlings under low oxygen and flooding stresses. Three-day-old soybean seedlings were subjected to low oxygen and flooding stresses. Growth of root was suppressed in both stresses with more extent of suppression in flooded seedlings at 3 and 6 days following the treatments. Proteins were extracted from roots and separated by two-dimensional polyacrylamide gel electrophoresis. Of total 1,233 protein spots, 27 protein spots were commonly changed under low oxygen and flooding stresses; while the differential change in 4 and 18 protein spots was specific to low oxygen and flooding stresses, respectively. Proteins related to metabolism and energy were increased; while protein destination/storage related proteins were decreased commonly under low oxygen and flooding stresses. Protein specie, TCP domain class transcription factor was decreased specifically under low oxygen stress; while decrease of nine proteins related to metabolism, protein destination/storage and disease/defense was specific in flooded seedlings. The decrease in majority of the proteins related to protein destination/storage specifically in flooded seedlings implies the misfolding of proteins resulting in flooded injuries in an independent way of oxygen deprivation. These results suggest that decrease in proteins related to protein destination/storage and disease/defense causes more growth suppression in soybean seedlings under flooding stress compared to low oxygen stress.





Similar content being viewed by others
Abbreviations
- 2-DE:
-
Two-dimensional polyacrylamide gel electrophoresis
- CBB:
-
Coomassie brilliant blue
- MS:
-
Mass spectrometry
- LC:
-
Liquid chromatography
- pI:
-
Isoelectric point
- IEF:
-
Isoelectric focusing
- MALDI-TOF:
-
Matrix-assisted laser desorption ionization time-of-flight
References
Gibbs J, Greenway H (2003) Mechanisms of anoxia tolerance in plants. I. Growth survival and anaerobic catabolism. Funct Plant Biol 30:1–47
Greenway H, Waters I, Newsome J (1992) Effects of anoxia on uptake and loss of solutes in roots of wheat. Aust J Plant Physiol 19:233–247
Rawyler A, Pavelic D, Gianinazzi C, Oberson J, Braendle R (1999) Membrane lipid integrity relies on a threshold of ATP production rate in potato cell cultures submitted to anoxia. Plant Physiol 120:293–300
Jackson MB, Colmer TD (2005) Response and adaptation by plants to flooding stress. Ann Bot 96:501–505
Huck MG (1970) Variation in tap root elongation as influenced by composition of soil air. Agron J 62:815–818
Crawford RMM (1992) Oxygen availability as an ecological limit to plant distribution. Adv Ecol Res 23:93–185
Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, Peacock WJ, Dolferus R, Dennis ES (2002) Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 14:2481–2494
Lasanthi-Kudahettige R, Magneschi L, Loreti E, Gonzali S, Licausi F, Novi G, Baretta O, Vitulli F, Alpi A, Perata P (2007) Transcript profiling of the anoxic rice coleoptiles. Plant Physiol 144:218–231
Komatsu S, Yamamoto R, Nanjo Y, Mikami Y, Yunokawa H, Sakata K (2009) Comprehensive analysis of the soybean genes and proteins expressed under flooding stress using transcriptome and proteome techniques. J Proteome Res 8:4766–4778
Nanjo Y, Maruyama K, Yasue H, Yamaguchi-Shinozaki K, Shinozaki K, Komatsu S (2011) Transcriptional responses to flooding stress in roots including hypocotyls of soybean seedlings. Plant Mol Biol 77:129–144
Russell DA, Wong DM-L, Sachs MM (1990) The anaerobic response of soybean. Plant Physiol 92:401–407
Shi F, Yamamoto R, Shimamura S, Hiraga S, Nakayama N, Nakamura T, Yukawa K, Hachinohe M, Matsumoto H, Komatsu S (2008) Cytosolic ascorbate peroxidase 2 (cAPX 2) is involved in the soybean response to flooding. Phytochemistry 69:1295–1303
Hashiguchi A, Sakata K, Komatsu S (2009) Proteome analysis of early-stage soybean seedlings under flooding stress. J Proteome Res 8:2058–2069
Nanjo Y, Skultety L, Ashraf Y, Komatsu S (2010) Comparative proteomic analysis of early-stage soybean seedlings responses to flooding by using gel and gel-free techniques. J Proteome Res 9:3989–4002
Drew MC, Lynch JM (1980) Soil anaerobiosis microorganisms and root function. Annu Rev Phytopathol 18:37–66
Boru G, Vantoai T, Alves J, Hua D, Knee M (2003) Responses of soybean to oxygen deficiency and elevated root-zone carbon dioxide concentration. Ann Bot 91:447–453
Komatsu S, Sugimoto T, Hoshino T, Nanjo Y, Furukawa K (2010) Identification of flooding stress responsible cascades in root and hypocotyl of soybean using proteome analysis. Amino Acids 38:729–738
Yanagawa Y, Komatsu S (2012) Ubiquitin/proteasome-mediated proteolysis is involved in the response to flooding stress in soybean roots independent of oxygen limitation. Plant Sci 185–186:250–258
Chen JJ, Chen CL, Sung FJM (1992) Flooding effect on photosynthesis in soybean leaves. J Agric For 41:17–26
Sallam A, Scott HD (1987) Effects of prolonged flooding on soybean at the R2 growth stage. I. Dry matter and N and P accumulation. J Plant Nutr 10:567–592
Minchin FR, Summerfield RJ (1976) Symbiotic nitrogen-fixation and vegetative growth of cowpea (Vigna unguiculata (L) Walp) in waterlogged conditions. Plant Soil 45:113–127
Sung FJM (1993) Waterlogging effects on nodule nitrogenase and leaf nitrate reductase activities in soybean. Field Crops Res 35:183–189
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang X-C, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkamp R, Dirkse W, Van Staveren M, Stiekema W (1998) Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391:485–488
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucl Acids Res 35:585–587
Huang S, Greenway H, Colmer TD, Millar AH (2005) Protein synthesis by rice coleoptiles during prolonged anoxia: implications for glycolysis growth and energy utilization. Ann Bot 96:661–668
Tan-Wilson AL, Rightmire BR, Wilson KA (1982) Different rates of metabolism of soybean proteinase inhibitors during germination. Plant Physiol 70:493–497
Richards KD, Schott EJ, Sharma YK, Davis KR, Gardner RC (1998) Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiol 116:409–418
Nanjo Y, Skultety L, Uvackova L, Klubicova K, Hajduch M, Komatsu S (2012) Mass spectrometry-based analysis of proteomic changes in the root tips of flooded soybean seedlings. J Proteome Res 11:372–385
Snowden KC, Richards KD, Gardner RC (1995) Aluminum induced genes. Induction by toxic metals, low Ca and wounding, and pattern of expression in root tips. Plant Physiol 107:341–347
Komatsu S, Deschamps T, Hiraga S, Kato M, Chiba M, Hashiguchi A, Tougou M, Shimamura S, Yasue H (2011) Characterization of a novel flooding stress responsive alcohol dehydrogenase expressed in soybean roots. Plant Mol Biol 77:309–322
Tenhaken R, Thulke O (1996) Cloning of an enzyme that synthesizes a key nucleotide- sugar precursor of hemicellulose biosynthesis from soybean: UDP-glucose dehydrogenase. Plant Physiol 112:1127–1134
Thapanapongworakul N, Nomura M, Van Dao T, Shimoda Y, Sato S, Tabata S, Tajima S (2010) 3-Phosphoglycerate dehydrogenase in Mesorhizobioum loti is essential for maintaining symbiotic nitrogen fixation of Lotus japonicus root nodules. Plant Soil 336:233–240
Gass N, Glagotskaia T, Mellema S, Stuurman J, Barone M, Mandel T, Roessner-Tunali U, Kuhlemeier C (2005) Pyruvate decarboxylase provides growing pollen tubes with a competitive advantage in petunia. Plant Cell 17:2355–2368
Gaikwad A, Long DJ 2nd, Stringer JL, Jaiswal AK (2001) In vivo role of NAD(P)H: quinone oxidoreductase I (NQO I) in the regulation of intracellular redox state and accumulation abdominal adipose tissue. J Dial Chem 276(22559):22564
Knogge W, Schmelzer E, Weissenbock G (1986) The role of chalcone synthase in the regulation of flavonoid biosynthesis in developing oat primary leaves. Arch Biochem Biophys 25:364–372
Cubas P, Lauter N, Loebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18:215–222
Viola IL, Uberti Manassero NG, Ripoll R, Gonzalez DH (2011) The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem J 435:143–155
Casati P, Walbot V (2004) Crosslinking of ribosomal proteins to RNA in maize ribosomes by UV-B and its effects on translation. Plant Physiol 136:3319–3332
Bailey-Serres J, Vangala S, Szick K, Lee CH (1997) Acidic phosphoprotein complex of the 60S ribosomal subunit of maize seedling roots. Components and changes in response to flooding. Plant Physiol 114:1293–1305
Crowell DN, John ME, Russel D, Amasino RM (1992) Characterization of a stress-induced developmentally regulated gene family from soybean. Plant Mol Biol 18:459–466
Alkharouf N, Khan R, Matthews B (2004) Analysis of expressed sequence tags from roots of resistant soybean infected by the soybean cyst nematode. Genome 47:380–388
Liu S, Cheng Y, Zhang X, Guan Q, Nishiuchi S, Hase K, Takano T (2007) Expression of an NADP-malic enzyme gene in rice (Oryza sativa. L) is induced by environmental stresses; over-expression of the gene in Arabidopsis confers salt and osmotic stress tolerance. Plant Mol Biol 64:49–58
Asada K (1999) The water–water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Phys 50:601–640
Narendra S, Venkataramani S, Shen G, Wang J, Pasapula V, Lin Y, Kornyeyev D, Holaday AS, Zhang H (2006) The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. J Exp Bot 57:3033–3042
Akhtar TA, Orsomando G, Mehrshahi P, Lara-Núñez A, Bennett MJ, Gregory JF 3rd, Hanson AD (2010) A central role for gamma-glutamyl hydrolases in plant folate homeostasis. Plant J 64:256–266
Hanson AD, Roje S (2001) One-carbon metabolism in higher plants. Annu Rev Plant Physiol Plant Mol Biol 52:119–137
Rebeille F, Ravanel S, Jabrin S, Douce R, Storozhenko S, Van der Straeten D (2006) Folates in plants: biosynthesis, distribution, and enhancement. Physiol Plant 126:330–342
Kwade Z, Swiatek A, Azmi A, Goossens A, Inze D, Van Onckelen H, Roef L (2005) Identification of four adenosine kinase isoforms in tobacco by-2 cells and their putative role in the cell cycle-regulated cytokinin metabolism. J Biol Chem 280:17512–17519
Moffatt BA, Stevens YY, Allen MS, Snider JD, Pereira LA, Todorova MI, Summers PS, Weretilnyk EA, Martin-McCaffrey L, Wagner C (2002) Adenosine kinase deficiency is associated with developmental abnormalities and reduced transmethylation. Plant Physiol 128:812–821
Kamauchi S, Wadahama H, Iwasaki K, Nakamoto Y, Nishizawa K, Ishimoto M, Kawada T, Urada R (2008) Molecular cloning and characterization of two soybean protein disulfide isomerases as molecular chaperones for seed storage proteins. FEBS J 275:2644–2658
Komatsu S, Hiraga S, Yanagawa Y (2012) Proteomics technique for the development of flood tolerant crops. J Proteome Res 11:68–78
Acknowledgments
The authors thank to the students exchange programme between Kohat University of Science and Technology, Pakistan and University of Tsukuba, Japan, for providing scholarship. We are grateful to the National Institute of Crop Science of Japan for all experimental support during this project. Authors thank to Dr. Yohei Nanjo and Dr. Keito Nishizawa for their valuable discussion. This work was supported by the Grants from National Agriculture and Food Research Organization, Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khatoon, A., Rehman, S., Oh, MW. et al. Analysis of response mechanism in soybean under low oxygen and flooding stresses using gel-base proteomics technique. Mol Biol Rep 39, 10581–10594 (2012). https://doi.org/10.1007/s11033-012-1946-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-1946-8