Abstract
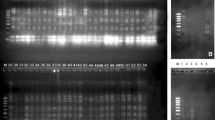
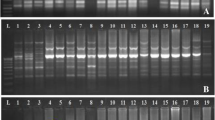
Fifty-two genotypes of Eleusine coracana collected from Uttarakhand hills were subjected to simple sequence repeat (SSR), random amplified polymorphic DNA (RAPD)-PCR and protein profiling analysis to investigate the variation in protein content. The main objective of the present study was to detect variability among E. coracana and also assess the discriminating ability of these three molecular methods. A total of 21 RAPD and 24 SSR primers were assayed for their specificity in detecting genetic variability in E. coracana, of which 20 RAPD and 21 SSR primers were highly reproducible and were found suitable for use in PCR analysis. Assessing genetic diversity among E. coracana genotypes by RAPD-PCR using 20 polymorphic primers yielded 56 different RAPD markers which clustered the genotypes into different groups on the basis of protein content. Similarly, SSR-PCR with 21 polymorphic primers clustered the genotypes into different groups. On the other hand, biochemical typing of E. coracana using whole seed proteins generated profiles that showed no major difference indicating the technique to be not useful in typing genotypes of this crop. However, a few of the genotypes showed the presence of a unique band of 32 kDa that needs to be further investigated to understand the role of the protein from nutritional point of view, if any. In the present study, significant negative correlation (r = −0.69*) was found between the protein and calcium content of finger millet genotypes. Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis based seed storage proteins generated profiles showed no major differences in banding pattern among 52 finger millet genotypes while quantitative estimation of seed storage protein fractions using Lowry method revealed that glutelin was highest followed by prolamin, globulin and albumin.




Similar content being viewed by others
References
Rachie KO, Peters LV (1977) The Eleusines: a review of the world literature. International Crops Research Institute for the Semi Arid Tropics (ICRISAT), Hyderabad, pp 118–136
Hilu KW, Johnson JL (1992) Ribosomal DNA hybridization in finger millet and wild species of Eleusine (Poaceae). Theor Appl Genet 83:895–902
Worth C, Sniegowski BP, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220
Muza SR, Fulco CS, Lyons T, Rock PB, Beidleman BA, Kenney J, Cymerman A (1995) Augmented chemosensitivity at altitude and after return to sea level: impact on subsequent return to altitude. Acta Andina 4:109–112
Salimath SS, Olivera ACD, Godwin ID, Bennetzen JL (1995) Assessment of genome origins and diversity in the genus Eleusine with DNA markers. Genome 38:757–763
Vadivoo AS, Joseph R, Ganesan NM (1998) Genetic variability and diversity for protein and calcium contents in finger millet Eleusine coracana (L. Gaertn.) in relation to grain color. Plant Foods Hum Nutr 52:353–364
Tingey SV, del Tufo JP (1993) Genetic analysis with random amplified polymorphic DNA markers. Plant Physiol 101:349–352
Mccouch SR, Tanksley SD (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Joshi SP, Gupta VS, Aggarwal RK, Ranjekar PK, Brar DS (2000) Genetic diversity and phylogenetic relationship as revealed by Inter simple sequence repeat polymorphism in the genus Oryza. Theor Appl Genet 100:1311–1320
Giovannoni JJ, Wing RA, Ganal MW, Tanksley S (1991) Isolation of molecular markers from specific chromosomal intervals using DNA pools from existing mapping populations. Nucl Acids Res 19:6553–6558
Cordeiro GM, Christopher MJ, Henry RJ, Reinke RF (2002) Identification of microsatellite markers for fragrance in rice by analysis of the rice genome sequence. Mol Breed 9(4):245–250
Fristsch P, Rieseberg LH (1995) The use of random amplified polymorphic DNA (RAPD) in conservation genetics. In: Smither TB, Wayne RK (eds) Molecular genetic approaches in conservation. Oxford University Press, Oxford, pp 54–73
Kjølner S, Såstad SM, Taberlet P, Brochmann C (2004) Amplified fragment length polymorphism versus random amplified polymorphic DNA markers: clonal diversity in Saxifraga cernua. Mol Ecol 13:81–86
Garcia AAF, Benchimol LL, Barbosa AMM, Geraldi IO, Souza CLJ, Souza AP (2004) Comparison of RAPD, RFLP, AFLP and SSR markers for diversity studies in tropical maize inbred lines. Genet Mol Biol 27:579–588
Singh AK, Sharma RK, Singh NK, Bansal KC, Koundal KR, Mohapatra T (2006) Genetic diversity in ber (Ziziphus spp.) as revealed by AFLP markers. J Hortic Sci Biotechnol 81:205–210
Selvi A, Nair NV, Noyer JL, Singh NK, Balasundaram N, Bansal KC, Koundal KR, Mohapatra T (2006) AFLP analysis of phonetic organization and genetic diversity in the sugarcane complex, Saccharum and Erianthus. Genet Resour Crop Evol 53:831–842
Lim W, Mudge KW, Weston LA (2007) Utilization of RAPD markers to assess genetic diversity of wild populations of North American Ginseng (Panax quinquefolius). Planta Med 73:71–76
Souframanien J, Gopalakrishna T (2004) A comparative analysis of genetic diversity in blackgram genotypes using RAPD and ISSR markers. Theor Appl Genet 109:1687–1693
Awasthi AK, Nagaraja GM, Naik GV, Kanginakudru S, Thangavelu K, Nagaraju J (2004) Genetic diversity and relationships in mulberry (genus Morus) as revealed by RAPD and ISSR marker assays. BMC Genet 5:1
Baranger A, Aubert G, Arnau G, Lainé AL, Deniot G, Potier J, Weinachter C, Lejeune-Hé naut I, Lallemand J, Burstin J (2004) Genetic diversity within Pisum sativum using proteinand PCR-based markers. Theor Appl Genet 108:1309–1321
Sica M, Gamba G, Montieri S, Gaudio L, Aceto S (2005) ISSR markers show differentiation among Italian populations of Asparagus acutifolius L. BMC Genet 6:17
Roy A, Bandyopadhyay A, Mahapatra AK, Ghosh SK, Singh NK, Bansal KC, Koundal KR, Mohapatra T (2006) Evaluation of genetic diversity in jute (Corchorus species) using STMS, ISSR and RAPD markers. Plant Breed 125:292–297
Singh A, Chaudhury A, Srivastava PS, Lakshmikumaran M (2002) Comparison of AFLP and SAMPL markers for assessment of intra-population genetic variation in Azadirachta indica A. Juss. Plant Sci 162:17–25
Negi MS, Sabharwal V, Bhat SR, Lakshmikumaran M (2004) Utility of AFLP markers for the assessment of genetic diversity within Brassica nigra germplasm. Plant Breed 123(1):13–16
Odat N, Jetschke G, Hellwig FH (2004) Genetic diversity of Ranunculus acris L. (Ranunculaceae) populations in relation to species diversity and habitat type in grassland communities. Mol Ecol 13(5):1251–1257
Bahulikar RA, Stanculescu D, Preston CA, Baldwin IT (2004) ISSR and AFLP analysis of the temporal and spatial population structure of the post-fire annual, Nicotiana attenuata, in SW Utah. BMC Ecol 4:12
Zhao J, Wang X, Deng B, Lou P, Wu J, Sun R, Xu Z, Vromans J, Koornneef M, Bonnema G (2005) Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor Appl Genet 110:1301–1314
Shan F, Clarke HC, Plummer JA, Yan G, Siddique KHM (2005) Geographical patterns of genetic variation in the world collections of wild annual Cicer characterized by amplified fragment length polymorphisms. Theor Appl Genet 110:381–391
Menkir A, Kling JG, Badu-Apraku B, Ingelbrecht I (2005) Molecular marker-based genetic diversity assessment of Striga-resistant maize inbred lines. Theor Appl Genet 110:1145–1153
Wu YQ, Taliaferro CM, Bai GH, Martin DL, Anderson JA, Anderson MP, Edwards RM (2006) Genetic analyses of Chinese Cynodon accessions by flow cytometry and AFLP markers. Crop Sci 46:917–926
Ru Z, Zhou C, Weifeng L, Baorong L (2006) Estimating genetic diversity and sampling strategy for a wild soybean (Glycine soja) population based on different molecular markers. Chin Sci Bull 51:1219–1227
Liu Y, Wang Y, Huang H (2006) High interpopulation genetic differentiation and unidirectional linear migration patterns in Myricaria laxiflora (Tamaricaceae), an endemic riparian plant in the Three Gorges Valley of the Yangtze River. Am J Bot 93:206–215
Han J, Zhang W, Cao H, Chen S, Wang Y (2007) Genetic diversity and biogeography of the traditional Chinese medicine, Gardenia jasminoides, based on AFLP markers. Biochem Syst Ecol 35:138–145
Zhou M-Q, Zhao K-G, Chen L-Q (2007) Genetic diversity of Calycanthaceae accessions estimated using AFLP markers. Sci Hortic (in press)
Ghafoor A, Afzal M, Anwar R (2002) Diversity in food legumes for sustainable utilization of plant genetic resources. In: Anwar R, Takahashi J, Bhatti MS, Masood S (eds) Sustainable utilization of plant genetic resources. PARC/IPGRI/JICA, Islamabad, pp 238–250
Ladizinsky G, Hymowitz T (1979) Seed protein electrophoresis in taxonomic and evolutionary studies. Theor Appl Genet 54:145–151
Das S, Mukarjee KK (1995) Comparative study on seed proteins of Ipomoea. Seed Sci Technol 23:501–509
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4326
Maniatis T, Sambrook J, Fritsch EF (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaudoise Sci Nat 44:223–270
Rohlf FJ (1997) NTSYS-pc numerical taxonomy and multivariate analysis system, version 2.0. Exeter Publications, New York
Yap IV, Nelson RJ (1996) Winboot: a program for performing bootstrap analysis of binary data to determine the confidence of UPGMA-based dendrograms. IRRI, Manilla
Tatham AS et al (1996) Characterisation of the Major Prolamins of Tef (Eragrostis tef) and Finger Millet (Eleusine coracana). J Cereal Sci 24:65–71
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Panwar P, Saini RK, Sharma N, Yadav D, Kumar A (2010) Efficiency of RAPD, SSR, and Cytochrome P450 gene based marker in accessing genetic variability amongst finger millet (Eleusine coracana) accessions. Mol Biol Rep. doi:10.1007/s11033-010-0067-5
Ayyangar GNR (1934) Recent work on the genetics of millets in India. Madras Agric J 22:16–26
Adrian J, Jaquot P (1964) Le sorgho et les mils en alimentation humaine et animale. Centre de Recherches sur la Nutrition, Bellevue (Seine et Olse). Vigot Frères, Editeurs
Ramiah PV, Satyanarayana P (1936) Biological values of ragi proteins. Proc Soc Biol Chem 1:7–8
Sastri BN (1939) Ragi (Eleusine coracana L. Gaertn), a new ragi material for the malting industry. Curr Sci 8:34–35
Ghafoor A (2003) [Vigna mungo (L.) Hepper] Identification of early maturing and high yielding lines of black gram. Proc Pak Acad Sci 40:127–134
Piergiovanni AR, Taranto G (2003) Geographic distribution of genetic variation in a lentil collection as revealed by SDS-PAGE fractionation of seed storage proteins. J Genet Breed 57:39–46
Sultana T, Ghafoor A, Ashraf M (2006) Geographic patterns of diversity of cultivated lentil germplasm collected from Pakistan, as assessed by seed protein assays. Acta Biol Crac Ser Bot Pol 48:77–78
Javaid A, Ghafoor A, Anwar R (2004) Seed storage protein electrophoresis in groundnut for evaluating genetic diversity. Pak J Bot 36(1):25–29
Jha SS, Ohri D (1996) Phylogenetic relationships of Cajanus cajan (L.) Millsp. (pigeonpea) and its wild relatives based on seed protein profiles. Genet Res Crop Evol 43:275–281
First International Small Millet Workshop (1986). http://www.fsifee.u-akugei.ac.jp/millets/1stintsym.htm
Project All India Coordinated Small Millets Improvement (1996) Annual Report University of Agriculture Sciences. Bangalore, India
Watanabe KN, Iwanaga M (1999) Plant genetic resources and its global contribution. Plant Biotechnol 16:7–13
Evaluation of finger millet germplasm (1996) Project Report All India Coordinated Research Project, Gandhi Krishi Vigyan Kendra, University of Agricultural Sciences. Bangalore, India
Acknowledgments
The authors wish to acknowledge the Department of Biotechnology, Govt. of India for providing financial support in the form of Programme Support for research and development in Agricultural Biotechnology at G.B. Pant University of Agriculture and Technology, Pantnagar (Grant No. BT/PR7849/AGR/02/374/2006). The authors thank the, Ranichauri Hill Campus G.B. Pant University of Agriculture and Technology for providing the seed samples of the germplasm analyzed in present study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kumar, A., Sharma, N., Panwar, P. et al. Use of SSR, RAPD markers and protein profiles based analysis to differentiate Eleusine coracana genotypes differing in their protein content. Mol Biol Rep 39, 4949–4960 (2012). https://doi.org/10.1007/s11033-011-1291-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1291-3