Abstract
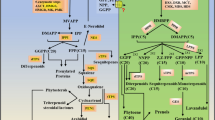
As a key enzyme in the biosynthesis of flavonols, anthocyanidins and proanthocyanidins, flavanone-3ß-hydroxylase (F3H) plays very important roles in plant stress response. A putative flavanone-3ß-hydroxylase gene from Medicago truncatula (MtF3H), a model legume species, was identified from a bio-data analysis platform. It was speculated to be induced by salt stress based on the outcomes of the analysis platform. The complementary DNA (cDNA) consists of 1499 bp with an open reading frame (ORF) of 1098 bp, which encodes a putative protein of 365 amino acids with a molecular weight of about 41.36 kDa and an isoelectric point of 5.60. To measure the catalytic activity of the protein, the MtF3H gene was ligated to pYES2 vector and heterologously expressed in yeast. The recombinant protein converted naringen into dihydrokaempferol and displayed different enzymatic efficiencies with other flavanones, confirming that MtF3H coding a functional flavanone-3ß-hydroxylase. The expression pattern of the MtF3H gene was analyzed by comparative quantitative RT-PCR and a higher level of expression was observed in the roots than was observed in stems and leaves. Furthermore, the expression was induced by salt stress in the roots, and to a greater extent in the stems, but the response of the gene activity to salt stress in the stems was slower in the first 12 h following treatment when compared to the roots.



Similar content being viewed by others
References
Tahara S (2007) A journey of twenty-five years through the ecological biochemistry of flavonoids. Biosci Biotech Bioch 71:1387–1404. doi:10.1271/bbb.70028
Dixon RA, Steele CL (1999) Flavonoids and isoflavonoids—a gold mine for metabolic engineering. Trends Plant Sci 4:394–400. doi:10.1016/S1360-1385(99)01471-5
Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55:481–504. doi:10.1016/S0031-9422(00)00235-1
Martens S, Mithöfer A (2005) Flavones and flavone synthases. Phytochemistry 66:2399–2407. doi:10.1016/j.phytochem.2005.07.013
Prescott AG, John P (1996) Di-oxygenases: molecular structure and role in plant metabolism. Annu Rev Plant Physiol Plant Mol Biol 47:245–271. doi:10.1146/annurev.arplant.47.1.245
Wellmann F, Matern U, Lukačin R (2004) Significance of C-terminal sequence elements for Petunia flavanone-3ß-hydroxylase activity. FEBS Lett 561:149–154. doi:10.1016/S0014-5793(04)00159-0
Britsch L, Ruhnau-Brich B, Forkmann G (1992) Molecular cloning, sequence analysis, and in vitro expression of flavanone-3ß-hydroxylase from Petunia hybrida. J Biol Chem 267:5380–5387
Davies KM (1993) A cDNA clone for flavanone 3-hydroxylase from Malus. Plant Physiol 103:291
Kim JH, Lee YJ, Kim BG, Lim Y, Ahn JH (2008) Flavanone-3ß-hydroxylases from rice: key enzymes for favonol and anthocyanin biosynthesis. Mol cells 25:312–316
Pelletier MK, Shirley BW (1996) Analysis of flavanone 3-hydroxylase in Arabidopsis thaliana seedlings (Coordinate regulation with chalcone synthase and chalcone isomerase). Plant Physiol 111:339–345
Jin Z, Grotewold E, Qu W, Fu G, Zhao D (2005) Cloning and characterization of a flavanone 3-hydroxylase gene from Saussurea medusa. DNA Seq 16:121–129
Shen G, Pang Y, Wu W, Deng Z, Zhao L, Cao Y, Sun X, Tang K (2006) Cloning and characterization of a flavanone 3-hydroxylase gene from Ginkgo biloba. Biosci Rep 26:19–29. doi:10.1007/s10540-006-9007-y
Charrier B, Coronado C, Kondorosi A, Ratet P (1995) Molecular characterization and expression of alfalfa (Medicago sativa L.) flavanone-3-hydroxylase and dihydroflavonol-4-reductase encoding genes. Plant Mol Biol 29:773–786
Baek MH, Chung BY, Kim JH, Wi SG, An BC, Kim JS, Lee SS, Lee IJ (2008) Molecular cloning and characterisation of the flavanone-3-hydroxylase gene from Korean black raspberry. J Hort Sci Biotech 83:595–602
Martens S, Forkmann G, Britsch L, Wellmann F, Matern U, Lukacin R (2003) Divergent evolution of flavonoid 2-oxoglutarate-dependent dioxygenases in parsley. FEBS Lett 544:93–98. doi:10.1016/S0014-5793(03)00479-4
Owens DK, Crosby KC, Runac J, Howard BA, Winkel BSJ (2008) Biochemical and genetic characterization of Arabidopsis thaliana flavanone-3ß-hydroxylase. Plant Physiol Biochem 46:833–843. doi:10.1016/j.plaphy.2008.06.004
Britsch L, Grisebach H (1986) Purification and characterization of (2S)-flavanone 3-hydroxylase from Petunia hybrida. Eur J Biochem/FEBS 156:569–577. doi:10.1111/j.1432-1033.1986.tb09616.x
Britsch L, Dedio J, Saedler H, Forkmann G (1993) Molecular characterization of flavanone-3ß-hydroxylases. Consensus sequence, comparison with related enzymes and the role of conserved histidine residues. Eur J Biochem/FEBS 217:745–754. doi:10.1111/j.1432-1033.1993.tb18301.x
Lukacin R, Groning I, Pieper U, Matern U (2000) Site-directed mutagenesis of the active site serine290 in flavanone-3ß-hydroxylase from Petunia hybrida. Euro J Biochem/FEBS 267:853–860. doi:10.1016/S0014-5793(00)01116-9
Lukacin R, Urbanke C, Groning I, Matern U (2000) The monomeric polypeptide comprises the functional flavanone-3ß-hydroxylase from Petunia hybrida. FEBS Lett 467:353–358. doi:10.1006/abbi.1999.1676
Lukacin R, Groning I, Schiltz E, Britsch L, Matern U (2000) Purification of recombinant flavanone-3ß-hydroxylase from Petunia Hybrida and assignment of the primary site of proteolytic degradation. Arch Biochem Biophys 375:364–370. doi:10.1046/j.1432-1327.2000.01064.x
Cook DR (1999) Medicago truncatula- a model in the making!. Curr Opin Plant Biol 2:301–304. doi:10.1016/S1369-5266(99)80053-3
Oldroyd GE, Geurts R (2001) Medicago truncatula, going where no plant has gone before. Trends Plant Sci 6:552–554. doi:10.1016/S1360-1385(01)02153-7
Young ND, Cannon SB, Sato S, Kim D, Cook DR, Town CD, Roe BA, Tabata S (2005) Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Physiol 137:1174–1181. doi:10.1104/pp.104.057034
Town CD (2006) Annotating the genome of Medicago truncatula. Curr Opin Plant Biol 9:122–127. doi:10.1016/j.pbi.2006.01.004
Sato S, Nakamura Y, Asamizu E, Isobe S, Tabata S (2007) Genome sequencing and genome resources in model legumes. Plant Physiol 144:588–593. doi:10.1104/pp.107.097493
Ane JM, Zhu H, Frugoli J (2008) Recent advances in Medicago truncatula genomics. Int J Plant Genomics 2008:256597 (Article ID). doi:10.1155/2008/256597
Mathesius U, Keijzer G, Natera SHA, Weinman JJ, Djordjevic MA, Rolfe BG (2001) Establishment of a root proteome reference map for the model legume Medicago truncatula using the expressed sequence tag database for peptide mass fingerprinting. Proteomics 1:1424–1440. doi:10.1002/1615-9861(200111)1:11<1424:AID-PROT1424>3.0.CO;2-J
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25:239–250. doi:10.1046/j.0016-8025.2001.00808.x
Munns R (2005) Genes and salt tolerance: bringing them together. New Phytol 167:645–663. doi:10.1111/j.1469-8137.2005.01487.x
Chinnusamy V, Jagendorf A, Zhu J-K (2005) Understanding and improving salt tolerance in plants. Crop Sci 45:437–448
Eryilmaz F (2006) The relationships between salt stress and anthocyanin content in higher plants. Biotechnol Biotechnol Equip 20:47–52
Oosten MV, Bressan R (2005) (abstract no. 518). The anthocyanin-impaired-response-1 (air-1) mutant in the sos3-1 background of Arabidopsis thaliana is deficient in the accumulation of anthocyanins in response to salt stress. 16th International Conference on Arabidopsis Research, www.arabidopsis.org/news/PosterAbstracts.pdf, pp 229
Barker DG, Pfaff T, Moreau D, Groves E, Ruffel S, Lepetit M, Whitehand S, Maillet F, Nair RM, Journet E-P (2006) Medicago truncatula handbook: Growing M. truncatula: choice of substrates and growth conditions, Available via DIALOG. http://www.noble.org/MedicagoHandbook/
Liu Y, Su Z, Dong J, Shen X, Li D, Wang T (2006) Construction and application of Medicago truncatula bio-data analysis platform. Acta Agrestia Sinica 14:231–235
Merchan F, de Lorenzo L, Rizzo SG, Niebel A, Manyani H, Frugier F, Sousa C, Crespi M (2007) Identification of regulatory pathways involved in the reacquisition of root growth after salt stress in Medicago truncatula. Plant J 51:1–17. doi:10.1111/j.1365-313X.2007.03117.x
de Lorenzo L, Merchan F, Blanchet S, Megías M, Frugier F, Crespi M, Sousa C (2007) Differential expression of the TFIIIA regulatory pathway in response to salt stress between Medicago truncatula genotypes. Plant Physiol 145:1521–1532. doi:10.1104/pp.107.106146
de Lorenzo L, Merchan F, Laporte P, Thompson R, Clarke J, Sousa C, Crespi M (2009) A novel plant leucine-rich repeat receptor kinase regulates the response of Medicago truncatula roots to salt stress. Plant Cell 21:668–680. doi:10.1105/tpc.108.059576
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔC(T) method. Methods 25:402–408. doi:10.1006/meth.2001.1262
Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32:327–331. doi:10.1093/nar/gkh454
Gebhardt YH, Witte S, Steuber H, Matern U, Martens S (2007) Evolution of flavone synthase I from parsley flavanone-3ß-hydroxylase by site-directed mutagenesis. Plant Physiol 144:1442–1454. doi:10.1104/pp.107.098392
Kim BG, Kim JH, Kim J, Lee C, Ahn JH (2008) Accumulation of flavonols in response to ultraviolet-B irradiation in soybean is related to induction of flavanone-3ß-hydroxylase and flavonol synthase. Mol cells 25:247–252
Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5:218–223. doi:10.1016/S1369-5266(02)00256-X
Ithal N, Reddy AR (2004) Rice flavonoid pathway genes, OsDfr and OsAns, are induced by dehydration, high salt and ABA, and contain stress responsive promoter elements that interact with the transcription activator, OsC1-MYB. Plant Sci 166:1505–1513. doi:10.1016/j.plantsci.2004.02.002
Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, Wanamaker SI, Mandal J, Xu J, Cui X, Close TJ (2005) Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol 139:822–835. doi:10.1104/pp.105.065961
Waller GR, Nowacki EK (1978) Alkaloid biology and metabolismin in plants. Plenum Press, New York, pp 121–141
Buer CS, Muday GK, Djordjevic MA (2007) Flavonoids are differentially taken up and transported long distances in Arabidopsis thaliana. Plant Physiol 145:478–490. doi:10.1104/pp.107.101824
Buer CS, Djordjevic MA (2009) Architectural phenotypes in the transparent testa mutants of Arabidopsis thaliana. J Exp Bot 60:751–763. doi:10.1093/jxb/ern323
Cho S, Chen W, Muehlbauer FJ (2005) Constitutive expression of the flavanone 3-hydroxylase gene related to pathotype-specific ascochyta blight resistance in Cicer arietinum L. Physiol Mol Plant Pathol 67:100–107. doi:10.1016/j.pmpp.2005.09.011
Acknowledgments
This work was supported by the Hi-Tech Research and Development (863) Program of China (2006AA10Z105, 2006AA100109). We thank Dr. Dasharath Lohar for providing seeds of M. truncatula Gaertn ‘Jemalong’ A17. We also sincerely thank Dr. shuizhang Fei (Iowa state University, Ames, USA) for his help.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shen, X., Martens, S., Chen, M. et al. Cloning and characterization of a functional flavanone-3ß-hydroxylase gene from Medicago truncatula . Mol Biol Rep 37, 3283–3289 (2010). https://doi.org/10.1007/s11033-009-9913-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-009-9913-8