Abstract
Rice (Oryza sativa) blast disease causes great annual yield losses and quality reductions. Limited resistance genes have been identified. Here, a genome-wide association study, together with an RNA sequencing analysis, was performed to identify novel marker–trait associations. In total, 127 associations were identified, explaining an average of 29.77% of the phenotypic variation. Five chromosomal hotspots were obtained. Moreover, 25 pleiotropic associations were associated with more than two strains. In particular, the lead single nucleotide polymorphism, seq–rs4199, close to Pi5 and Pi56(t) on chromosome 9, was associated with five different strains. In addition, 2341 nonredundant candidate genes, including 45 disease resistance-related genes, were predicted in a 200-kb genomic region for these associations. One gene, LOC_Os12g23930, overlapped a previously reported candidate resistance gene, and its relative expression level significantly decreased after inoculation. Furthermore, 683 differentially expressed rice genes, including 282 up- and 401 downregulated genes, were identified using RNA sequencing. By integrating our analyses techniques, three overlapping candidate genes, LOC_Os02g02850, LOC_Os02g41590, and LOC_Os02g41630, were obtained. The relative expression level of the former significantly increased after fungal inoculations, while the levels of the latter two significantly decreased. This work will help elucidate the molecular mechanisms of rice blast resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa) blast disease, caused by the fungus Magnaporthe oryzae (Bourett and Howard 1990), which is a teleomorph of the complex Ascomycete genus that is composed of interfertile anamorphs (Barr 1977; Dean et al. 2005), is a major constraint on rice yield and quality in the rice-planting regions of the world. The disease is difficult to combat owing to the ability of the causal fungal agent to adapt to its host and to frequently mutate. The long-term use of traditional chemical pesticides to control rice blast disease can lead to environmental pollution and is not economical. Thus, breeding disease-resistant cultivars is a more environmentally friendly and effective strategy to manage blast disease (Savary et al. 2012). The identification of resistance genes/quantitative trait loci (QTLs) and understanding disease-resistant mechanisms are particularly important for the breeding of disease-resistant plants. In rice, 216 blast resistance-related genes/QTLs have been identified (http://www.ricedata.cn/ontology, TO:0000074). On the basis of differences in conserved protein motifs, such as nucleotide-binding sites, leucine-rich repeats, Toll–interleukin receptors, coiled coils, transmembrane receptors, and protein kinases, resistance genes can be classified into different categories (Liu et al. 2007a). More than 100 resistance genes have been identified and at least 24 of these resistance genes/QTLs have been cloned (http://www.ricedata.cn/gene/gene_pi.htm).
Although most of the above resistance genes/QTLs were identified using linkage mapping (Wang et al. 1999; Lin et al. 2007; Takahashi et al. 2010; Zeng et al. 2011; Liu et al. 2013; Chen et al. 2015; Li et al. 2015), the detection of genes/QTLs is limited by the biparental materials used. With the development of high-density genetic mapping, genome-wide association studies (GWASs) based on linkage disequilibrium have become effective gene/QTL mapping strategies for natural populations (Zhu et al. 2008). GWASs overcome the shortcomings of linkage mapping that are associated with using a population derived from biparental materials (Yu et al. 2006). A series of GWASs have been successfully performed to dissect complex traits in multiple crop species, including maize (Riedelsheimer et al. 2012), soybean (Wen et al. 2018), wheat (Sukumaran et al. 2015), cotton (Du et al. 2018), and sorghum (Chopra et al. 2017). In rice, a high-density single nucleotide polymorphism (SNP) map and comprehensive HapMap were constructed several years ago (Huang et al. 2010). Subsequently, GWASs were successfully applied to uncover 14 complex rice agronomic traits, and 37 associations were identified (Huang et al. 2010). One of these associations, located on chromosome 7, was analyzed in depth, resulting in the identification of the candidate gene OsSPL13 that positively regulates cell size and enhances rice grains’ length and yield (Si et al. 2016). In addition, based on the same high-density SNP map and population (Huang et al. 2010), a GWAS was successfully applied to detect QTLs related to rice blast resistance, and 30 associated loci were identified (Wang et al. 2014).
Previously, we developed a custom-designed array (Lu et al. 2015) that consisted of 5291 SNPs evenly chosen from the Rice Haplotype Map Project Database (http://www.ncgr.ac.cn/ricehap2/) (Huang et al. 2010). We have successfully used the array to perform GWASs for different agronomic traits and identified multiple important associations that will be beneficial in the molecular breeding of rice in future (Lu et al. 2015, 2016, 2018; Feng et al. 2016; Zhang et al. 2017). Here, we examined rice blast resistance using a GWAS based on the custom-designed SNP array in 355 diverse indica accessions. The objectives of the present study were as follows: (1) to analyze the genetic architecture of rice blast resistance, (2) to detect a set of lead associations for candidate gene identification, and (3) to preliminarily verify novel functional candidate genes that might participant in regulating blast resistance.
Material and methods
Plant material and phenotypic evaluation
The association mapping panel was composed of 355 indica accessions that were selected from our previous study, which were described in detail by Lu et al. (2015). The phenotypic evaluation was performed at the China National Rice Research Institute in Hangzhou (N 30° 32′, E 120° 12′), China, in 2016. Seeds were planted in plastic trays (length × width × height, 43 × 30 × 7.5 cm) to test rice blast resistance using 16 M. oryzae strains collected from South China. Each accession was inoculated at the third to fourth leaf stage using the spray method and then planted in a greenhouse having a high temperature (~ 35 °C) and high humidity (~ 80%) environment. The disease level was evaluated 7 days after inoculation. The diseased leaf area (DLA) was used as the evaluation criterion (Chuwa et al. 2015): 0 = no lesions; 1 = small, brown, specks of pinhead size; 3 = small, roundish to slightly elongated, necrotic, gray spots about 1–2 mm in diameter; 5 = typical blast lesions infecting <10% of the leaf area; 7 = typical blast lesions infecting 26–50% of the leaf area; and 9 = typical blast lesions infecting > 51% leaf area and many dead leaves. Three replications were performed for each experiment.
Genotyping, population structure and association mapping
The genotypes of all of the accessions were obtained using a custom-designed array containing 5291 SNP markers reported in our previous study (Lu et al. 2015). The SNPs having minor allele frequencies < 5% were removed, resulting in 4032 SNPs being used in the association analyses.
The population structure analysis was performed using STRUCTURE v2.2 (Pritchard et al. 2000a) with the following parameters: 1 to 15 populations with five runs, 10,000 burn-in period, and 100,000 Markov Chain Monte Carlo replications. The principal component analysis was performed using PowerMarker v3.25 (Liu and Muse 2005) and NTSYSpc v2.1 (Rohlf 2000). The pairwise relatedness coefficients were estimated using SPAGeDi v 1.4c (Hardy and Vekemans 2002).
The association mapping was performed by TASSEL v4.0 (Bradbury et al. 2007), and the EMMA (Kang et al. 2008) and P3D (Zhang et al. 2010) algorithms were used to reduce the computing time. A mixed linear model, with the population structure (Q) and relative kinship matrix (K) as covariates, was used to control the rate of false positive associations. A compromised threshold of P = 1E−03 was used to identify the significant associations.
Candidate gene identification and expression profiling
The potential candidate genes near the associations were identified within a 200-kb genome region (± 100 kb of the lead SNPs). The annotations of these candidate genes were obtained from the Rice Haplotype Map Project Database (http://www.ncgr.ac.cn/ricehap2/). The relative expression levels of the target genes were assessed by quantitative real-time PCR (qRT-PCR). Total RNA was extracted from young leaves using a MiniBEST Plant RNA Extraction kit (TaKaRa Bio Inc., Japan), following the manufacturer’s instructions. Next, first-strand complementary DNA was synthesized using PrimeScript RT Master Mix (TaKaRa Bio Inc). Then, the reaction mixture was run on a 7500 Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA) (Lu et al. 2016). Rice Ubq-2 was used as the internal control, and all of the reactions were repeated using three independent biological replicates.
RNA sequencing
The blast resistance rice variety Shuidaobawang (CH491) was inoculated with the M. oryzae S182 strain. The uninoculated variety acted as the control. After lesions showed, total RNA was extracted from young leaves using a MiniBEST Plant RNA Extraction kit (TaKaRa Bio Inc) according to the instructions. Three biological replications for each treatment were used for transcriptome sequencing. The enrichment of mRNA, fragment interruption, addition of adapters, size selection, and PCR amplification were performed by CapitalBio Technology Ltd. (Beijing, China). The library was sequenced using the Illumina HiSeq™ 2500 platform.
Identification of differentially expressed genes
The raw sequencing data was collected, and low-quality reads, adaptor sequences, and empty reads were removed. Next, the transcriptomes were remapped to the reference genome (Oryza sativa L. spp. Japonica, var. Nipponbare, MSU 6.0) using HISAT to check the sequencing quality (Kim et al. 2015). The gene expression level was quantified as the total number of reads for each gene that uniquely aligned to the reference genome. To identify genes related to blast resistance, we selected genes that had expression levels that were significantly altered by treatment (inoculation) compared with the control. A false discovery rate ≤ 0.05 and an absolute value of log2 ratio ≥ 3 were used as the thresholds to judge the significance of differences in gene expression.
Results
Material distribution and phenotypic variation
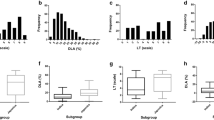
A set of 355 genetically diverse indica accessions collected from the 17 main provinces in South China, which represented the major rice-planting region, was used to perform the GWAS to detect novel loci associated with resistance to rice blast disease (Figure S1a; Table S1). A total of 16 strains of rice blast collected from eight provinces in South China were used to investigate blast resistance at the seedling stage (Figure S1a; Table 1; Table S2). The resistance level of each accession was evaluated by DLA after inoculation. Extensive and rich phenotypic variations were observed in the resistance to the 16 blast strains (Figure S1b). The disease rating as assessed by the DLA ranged from 2.99 for ‘S254’ to 6.68 for ‘S171’, with an average of 5.23 (Table 1).
The correlation analyses between disease rating and both longitude and latitude indicated that the DLA resulting from inoculations with the 16 strains was significantly positively correlated with latitude except for S102, while the DLAs caused by ‘S172’ (r = −0.12, p = 0.023), ‘S193’ (r = −0.15, p = 0.005), and ‘S366’ (r = −0.14, p = 0.011) were highly negatively associated with longitude (Table S3; Figure S2). Moreover, most of the correlation coefficients of disease rating as assessed by DLA resulting from inoculations of the 16 strains were also significantly (p < 0.05) or highly significantly (p < 0.01) positively correlated with each other, especially between ‘S122’ and both ‘S149’ (r = 0.62, p < 0.01) and ‘S366’ (r = 0.61, p < 0.01), while only the correlations between ‘S172’ and both ‘S182’ (r = 0.05) and ‘S254’ (r = 0.07) were not significant (p > 0.05) (Figure S3).
Population divergence and relative kinship analysis
Our previous study showed that the indica population can be classed into four subpopulations with moderate levels of differentiation (Lu et al. 2015). Here, we reused 355 indica accessions from the previous study as the GWAS population and reanalyzed the genetic component of each accession using STRUCTURE v2.2 software (Pritchard et al. 2000a). The maximum value of Evanno’s ΔK was K = 5 (Fig. 1a), suggesting that the genetic structure of the 355 indica accessions had five classifications (Fig. 1b), which was supported by the principal component analysis plot (Fig. 1c). The percentage of phenotypic variation explained by population structure ranged from 6.01 to 26.36%, with an average of 15.44% (Table 1). Thus, population structure might cause false positive associations that should not be ignored in further GWAS analyses (Yu et al. 2006). Most of the pairwise kinship coefficients were less than 0.1, indicating that there was no or weak relatedness among the accessions (Figure S4).
GWAS for resistance to 16 blast-causing M. oryzae strains
Our previous work with the indica panel suggested that false positive associates were only greatly controlled by a Q + K model (Lu et al. 2015). Thus, a GWAS for resistance to rice blast was performed to identify the associations using TASSEL v4.0 under the Q + K association model (Bradbury et al. 2007). To select the major associations and mined known loci, the lead associations with the lowest p values were maintained in a 200 (± 100)-kb region. After clumping, 127 significant associations, including eight known loci, were identified (Table 2; Table S4). All of the significant associations explained 29.77% of the average phenotypic variation, ranging from 15.59% for resistance to ‘S122’ to 49.50% for resistance to ‘S182’ (Table 2).
Most of the lead SNPs were very close to known genes. In rice varieties resistant to ‘S242’, Pi9 was only ~ 31 kb away from the significant association, seq–rs2897 (p = 7.10E−04) on chromosome 6, and in rice varieties resistant to ‘S172’, Pita was only ~ 64 kb away from lead SNP seq–rs5659 (p = 4.96E−08) on chromosome 12 and Pi1 was only ~ 89 kb away from the lead SNP seq–rs5411 (p = 5.92E−04) on chromosome 11 (Figure S5; Table S4). In addition, some associations were also identified in our previous GWAS repots using the different indica association population (Wang et al. 2014). For example, the two significant SNPs, seq–rs5460 (p = 8.54E−05) and seq–rs5489 (p = 9.00E−04), on chromosome 11 were associated with resistance to ‘S168’, and the lead SNP seq–rs3576 (p = 1.55E−06) on chromosome 7 was associated with resistance to ‘S366’ (Figure S5; Table S4). A set of significant associations was newly identified. For resistance to ‘S122’, a new lead SNP (seq–rs2233, p = 7.97E−04) and a cluster of new significant associations were detected on chromosomes 4 and 12, respectively. For resistance to ‘S182’, a new lead SNP (seq–rs5745, p = 1.09E−11), with the lowest p value, was obtained on chromosome 12. Moreover, for resistance to ‘S359’, a series of significant associations lead by seq–rs2863 (p = 4.19E−05) was detected on chromosome 6 (Figure S5; Table S4).
Assessment of GWAS significant associations
The greatest number of associations, accounting for 34.6% of all the associations, was identified on chromosome 12. In addition, 13.4% of all associations was distributed on chromosome 9, followed by chromosomes 11 (7.9%) and 6 (10.2%) (Fig. 2a). Furthermore, of the associations distributed on these four chromosomes, a series of hotspots was observed (Fig. 2b). In rice, chromosomal hotspots for rice blast resistance were frequently identified, especially on chromosomes 6, 11, and 12 (http://www.ricedata.cn/gene).
Further analyses of these associations revealed that 25 associations, including four known genes, showed pleiotropy that was associated with more than two strains (Fig. 2c). For example, the lead SNP (seq–rs4199, Pos: ~ 10.35 Mb) located on chromosome 9 was associated with five different strains, which was supported by the correlation coefficient value among the DLAs caused by the five strains (Figure S3). Moreover, the lead SNP is 117.25 kb and 41.94 kb away from two known blast resistance genes, Pi5 and Pi 56(t), respectively (Lee et al. 2009; Liu et al. 2013). Thus, the two genes appear to be important candidate genes for broad-spectrum resistance to blast, as previously reported (Liu et al. 2013). In addition, two other broad-spectrum resistance genes, Pi9 (Qu et al. 2006) and Pita (Bryan et al. 2000; Tacconi et al. 2010), were also obtained (Fig. 2c). Apart from the three known loci, 22 new pleiotropic associations were identified, including seq–rs4185 on chromosome 9 and seq–rs5745 on chromosome 12, which were correlated in four different strains (Fig. 2c). These pleiotropic associations would be very helpful in the molecular breeding of blast resistance in rice.
The efficacy of pyramiding elite alleles for different numbers of associations and random markers was examined to determine their effects in different strains, without considering the interaction among the associations/markers. The rice blast resistance increased (disease rating decreased) with the pyramiding of greater numbers of elite alleles (Figure S6a). However, the resistance level to rice blast remained almost unchanged, even as more elite alleles of random markers were pyramided (Figure S6b). Thus, enhancing the frequencies of elite alleles of significant associations could increase rice blast resistance.
Identification of candidate genes for the associations
For the 127 associations, 3877 candidate genes were identified within a 200-kb genomic region for each association from the Rice Haplotype Map Project Database (http://www.ncgr.ac.cn/ricehap2/). After clumping, 2341 nonredundant candidate genes were obtained (Table S5), including 45 disease resistance-related genes (Table S6). Of these 45 disease resistance candidate genes, 13 had been previously identified (Lopez–Gerena 2006; Shi et al. 2011; Jung et al. 2014; Kang et al. 2016), five were cloned in previous studies, such as Pi9 (LOC_Os06g17900), Pid2 (LOC_Os06g29810), Pi5 (LOC_Os09g15840), Pi56(t) (LOC_Os09g16000), and Pita (LOC_Os12g18360), and 1 (LOC_Os12g23930) overlapped with our previous GWAS results using a different population (Wang et al. 2014) (Figure S7a; Table S6). The 45 resistance-related genes were mainly distributed on chromosome 11, followed by chromosomes 6 and 8 (Figure S7b). This was consistent with the clustering distribution of the associations (Fig. 2a, b). Furthermore, the four lead SNPs, seq–rs2897, seq–rs5460, gwseq–rs12, and seq–rs722, were identified in a relatively large number of resistant genes (Figure S7c), indicating that they would be helpful in breeding a rice variety with broad-spectrum and high-level resistance to rice blast. The relative expression level of the candidate gene LOC_Os12g23930 (Os12g0427000), which was also identified in our previous study, highly significantly decreased after inoculation (Figure S7d; Table S7). Its relative expression pattern was consistent with our previous report (Wang et al. 2014), suggesting that the candidate gene might be involved in negatively regulating rice blast resistance.
Identification of blast resistance genes by combining GWAS and RNA sequencing
To identify blast resistance candidate genes, we inoculated a medium-resistance rice variety (CH491) with the M. oryzae S182 strain. ‘CH491’ that was not inoculated acted as the control. After the lesions appeared, RNA from the leaves was sequenced (unpublished). A total of 232,180,474 reads were generated using the Illumina sequencing platform. Of these reads, a set of 91,869,902 reads were mapped (Table S8). In total, 683 significantly differentially expressed genes, including 282 up- and 401 downregulated genes, were identified (Table S9; Fig. 3a). Among these differentially expressed candidate genes, eight genes were functionally annotated as encoding resistance proteins (Table S10). In addition, one of the eight genes, LOC_Os01g66020, was presumed to be associated with rice leaf blight resistance in a previous report (Jung et al. 2014).
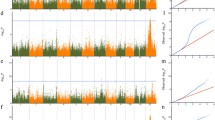
Candidate gene identification by integrating GWAS and RNA sequencing analyses. a Detection of differentially expressed genes by RNA sequencing. Blue and red crosses represent up- and downregulated genes, respectively. Green crosses represent unchanged genes. b Association peaks of the three overlapped genes with two lead SNPs. c Three overlapped genes identified by RNA sequencing and GWAS for S182 strain. d Relative expression levels of the three target genes. Error bar is standard error. One and double asterisks represent t test at 0.05 and 0.01 significant level, respectively
For resistance to ‘S182’, 382 blast resistance candidate genes were detected by the GWAS analyses (Fig. 3b; Table S9). Three, LOC_Os02g02850, LOC_Os02g41590, and LOC_Os02g41630, were confirmed by the RNA sequencing analyses (Fig. 3c; Table S10). The former is located near the association SNP seq–rs722 (p = 1.88E−04), and the latter two are close to the association SNP seq–rs1136 (p = 4.82E−04) on chromosome 2 (Fig. 3c; Table S5). A previous study suggested that the gene, LOC_Os02g41630, was a member of the phenylalanine ammonia-lyase gene family (OsPAL1), and it was highly homologous with the member OsPAL5. Furthermore, another member, OsPAL4, is a broad-spectrum disease resistance-related gene, and the resistance levels to three diseases, bacterial wilt, sheath blight, and rice blast, were significantly enhanced in heterozygous mutant materials (Tonnessen et al. 2015). Highly significant (p < 0.01) or significant (p < 0.05) expression level differences of the three candidate genes occurred before and after inoculation (Fig. 3d; Table S7).
Discussion
Here, a GWAS was used to identify associations of genes to rice blast resistance. Although GWAS is a useful strategy to reveal complicated traits and mine candidate genes (Huang et al. 2010; Wang et al. 2014; Lu et al. 2015, 2016, 2018; Si et al. 2016; Feng et al. 2016; Chopra et al. 2017; Zhang et al. 2017), population structure appears to affect the associations, which may result in false positive associations (Atwell et al. 2010). To eliminate false positive associations, structural association and genomic control have always been used in previous studies (Pritchard et al. 2000b; Yu et al. 2006). In our work, the accessions were selected from a previously reported indica-only panel (Lu et al. 2015), which should reduce false positive assertions. In addition, the population structure was reanalyzed using the program STRUCTURE v2.2 (Pritchard et al. 2000a), and the familial relationships were re-estimated using SPAGeDi (Hardy and Vekemans 2002). The panel contains no or weak relatedness among the accessions (Fig. 1; Figure S4). Previously, four GWAS models were used to evaluate the best association model in two environments, and the Q + K model was selected (Lu et al. 2015). Therefore, we used the Q + K model for the association mapping.
Here, we identified 127 associations for rice blast resistance, including eight known loci, such as Pita (Bryan et al. 2000; Tacconi et al. 2010), Pi9 (Qu et al. 2006), Pi5 (Lee et al. 2009), Pi56(t) (Liu et al. 2013), and Pid2 (Chen et al. 2006), in the indica-only panel. More importantly, a set of new significant associations with resistance to the 16 strains, especially S102, S122, S182, and S359 on chromosomes 6, 12, 12, and 6, respectively, was identified on 12 chromosomes (Figure S5; Table S4). In addition, we identified two associations with resistance to ‘S168’ on chromosome 7 and one lead SNP associated with resistance to ‘S366’ on chromosome 11. These had also been identified in our previous GWAS using a different association population (Wang et al. 2014). These new significant associations will aid in molecular breeding and functional candidate gene identification in the future.
Chromosomal hotspots have previously been identified for rice blast disease. For example, nine, 17, and 15 loci containing 15, 24, and 17 major blast resistance genes, respectively, were obtained on chromosomes 6, 11, and 12, respectively (http://www.ricedata.cn/gene). In our study, the associations also showed the presence of chromosomal hotspots on chromosomes 6, 9, 11, and 12, which was basically consistent with previous reports (Fig. 2b). Moreover, several lead SNPs were identified as being associated with the multiple strains (Fig. 2c), which might represent a multigenic effect, suggesting that a broad-spectrum resistance gene is near to the lead SNP. These candidate genes may share a common mechanism against different strains because of the pleiotropic or hitchhiking effect (Wang et al. 2014; Lu et al. 2015). In addition, these lead SNPs with pleiotropic and multigetic effects would be helpful for pyramid breeding in future.
To perform the GWAS, we used a custom-designed SNP array containing 5291 SNPs for the indica panel. Although, some GWASs involve ten thousand or a million SNP markers (Huang et al. 2010; Kang et al. 2016), given the linkage disequilibrium distance in the indica panel (~ 123 kb) (Huang et al. 2010), the genome coverage should be sufficient to identify most of the associations related to rice blast resistance. However, a higher density of SNPs would be helpful to identify causative genes and polymorphisms. For instance, the GWAS mapping resolution was improved for the target zone, from 220 to 20 kb, when using a 700-K SNP dataset rather than a 44-K SNP dataset (Kang et al. 2016).
Furthermore, we combined RNA sequencing and a GWAS analysis to identify blast resistance candidate genes against ‘S182’, and three overlapping genes were obtained (Fig. 3). One of the three genes, LOC_Os02g41630, is a member of phenylalanine ammonia-lyase gene family (OsPAL1), and another member, OsPAL4, is a broad-spectrum disease resistance-related gene for bacterial wilt, sheath blight, and rice blast (Tonnessen et al. 2015). The combined gene annotation and qRT-PCR were used to investigate the potential biological functions of the candidate gene. The gene’s expression level decreased after inoculation, which indicated that it may be involved in the negative regulation of rice blast resistance. Despite the relative expression verification, the biological functions of all of the candidate genes need to be validated using biotechnology experiments.
In summary, we identified a set of marker–trait associations and obtained multiple candidate genes for rice blast resistance using GWAS strategy. In addition, we combined a GWAS and RNA sequencing to mine for blast resistance candidate genes and obtained three overlapping candidate genes, which were further supported by the qRT-PCR analyses. Our report further confirms that GWAS is a powerful approach for discovering the quantitative blast resistance candidate genes compared with traditional linkage mapping, especially when used in combination with other analyses. This work provides a basis for further studies of the potential functions of these candidate genes. Our further work will also focus on validating the functional variants using molecular biology experiments. These broad-spectrum associations will be used to develop breeding markers for molecular assisted selection.
References
Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, Jiang R, Muliyati NW, Zhang X, Amer MA, Baxter I, Brachi B, Chory J, Dean C, Debieu M, de Meaux J, Ecker JR, Faure N, Kniskern JM, Jones JDG, Michael T, Nemri A, Roux F, Salt DE, Tang C, Todesco M (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465:627–631
Barr ME (1977) Magnaporthe, Telimenella, and Hyponectria (Physosporellaceae). Mycologia 69:952–966
Bourett TM, Howard RJ (1990) In vitro development of penetration structures in the rice blast fungus Magnaporthe grisea. Can J Bot 68:329–342
Bradbury PJ, Zhang ZW, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Bryan GT, Wu KS, Farrall L, Jia YL, Hershey HP, McAdams SA, Faulk KN, Donaldson GK, Tarchini R, Valent B (2000) A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12:2033–2046
Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Cao G, Ma B, Wang Y, Zhao X, Li S, Zhu L (2006) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J 46:794–804
Chen J, Peng P, Tian J, He Y, Zhang L, Liu Z, Yin D, Zhang Z (2015) Pike, a rice blast resistance allele consisting of two adjacent NBS–LRR genes, was identified as a novel allele at the Pik locus. Mol Breed 35:117
Chopra R, Burow G, Burke JJ, Gladman N, Xin Z (2017) Genome-wide association analysis of seedling traits in diverse Sorghum germplasm under thermal stress. BMC Plant Biol 17:12
Chuwa CJ, Mabagala RB, Reuben M (2015) Assessment of grain yield losses caused by rice blast disease in major rice growing areas in Tanzania. Int J Sci Res 4:2211–2218
Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan H, Read ND, Lee YH, Carbone I, Brown D, Oh YY, Donofrio N, Jeong JS, Soanes DM, Djonovic S, Kolomiets E, Rehmeyer C, Li W, Harding M, Kim S, Lebrun MH, Bohnert H, Coughlan S, Butler J, Calvo S, Ma LJ, Nicol R, Purcell S, Nusbaum C, Galagan JE, Birren BW (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980–986
Du X, Huang G, He S, Yang Z, Sun G, Ma X, Li N, Zhang X, Sun J, Liu M, Jia Y, Pan Z, Gong W, Liu Z, Zhu H, Ma L, Liu F, Yang D, Wang F, Fan W, Gong Q, Peng Z, Wang L, Wang X, Xu S, Shang H, Lu C, Zheng H, Huang S, Lin T, Zhu Y, Li F (2018) Resequencing of 243 diploid cotton accessions based on an updated a genome identifies the genetic basis of key agronomic traits. Nat Genet 50:796–802
Feng Y, Lu Q, Zhai R, Zhang M, Xu Q, Yang Y, Wang S, Yuan X, Yu H, Wang Y, Wei X (2016) Genome wide association mapping for grain shape traits in indica rice. Planta 244:819–830
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hua L, Wu J, Chen C, Wu W, He X, Lin F, Wang L, Ashikawa I, Matsumoto T, Wang L, Pan Q (2012) The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor Appl Genet 125:1047–1055
Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, Li M, Fan D, Guo Y, Wang A, Wang L, Deng L, Li W, Lu Y, Weng Q, Liu K, Huang T, Zhou T, Jing Y, Li W, Lin Z, Buckler ES, Qian Q, Zhang QF, Li J, Han B (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42:961–967
Jung KH, Han M, Nguyen VNT, Yoo Y, Nguyen MP, Lee C, Wang GL, Jeon JS (2014) Development of defense signaling pathways against bacterial blight disease in rice using genom-wide transcriptome data. J Agric Sci 6:48–111
Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E (2008) Efficient control of population structure in model organism association mapping. Genetics 178:1709–1723
Kang H, Wang Y, Peng S, Zhang Y, Xiao Y, Wang D, Qu S, Li Z, Yan S, Wang Z, Liu W, Ning Y, Korniliev P, Leung H, Mezey J, McCouch SR, Wang GL (2016) Dissection of the genetic architecture of rice resistance to the blast fungus Magnaporthe oryzae. Mol Plant Pathol 17:959–972
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
Lee SK, Song MY, Seo YS, Kim HK, Ko S, Cao PJ, Suh JP, Yi G, Roh JH, Lee S, An G, Hahn TR, Wang GL, Ronald P, Jeon JS (2009) Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding-leucine-rich repeat genes. Genetics 181:1627–1638
Li JB, Sun YD, Liu H, Wang YY, Jia YL, Xu MH (2015) Natural variation of rice blast resistance gene Pi-d2. Genet Mol Res 14:1235–1249
Lin F, Chen S, Que Z, Wang L, Liu X, Pan Q (2007) The blast resistance gene Pi37 encodes a nucleotide binding site leucine-rich repeat protein and is a member of a resistance gene cluster on rice chromosome 1. Genetics 177:1871–1880
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Liu J, Liu X, Dai L, Wang G (2007a) Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J Genet Genomics 34:765–776
Liu X, Lin F, Wang L, Pan Q (2007b) The in silico map-based cloning of Pi36, a rice coiled-coil nucleotide-binding site leucine-rich repeat gene that confers race-specific resistance to the blast fungus. Genetics 176:2541–2549
Liu Y, Liu B, Zhu X, Yang J, Bordeos A, Wang G, Leach JE, Leung H (2013) Fine-mapping and molecular marker development for Pi56(t), a NBS-LRR gene conferring broad-spectrum resistance to Magnaporthe oryzae in rice. Theor Appl Genet 126:985–998
Lopez–Gerena J (2006) Mapping QTL controlling durable resistance to rice blast in the cultivar Oryzica Llanos 5, Kansas State University, (Doctoral dissertation)
Lu Q, Zhang M, Niu X, Wang S, Xu Q, Feng Y, Wang C, Deng H, Yuan X, Yu H, Wang Y, Wei X (2015) Genetic variation and association mapping for 12 agronomic traits in indica rice. BMC Genomics 16:1067
Lu Q, Zhang M, Niu X, Wang C, Xu Q, Feng Y, Wang S, Yuan X, Yu H, Wang Y, Wei X (2016) Uncovering novel loci for mesocotyl elongation and shoot length in indica rice through genome-wide association mapping. Planta 243:645–657
Lu Q, Niu X, Zhang M, Wang C, Xu Q, Feng Y, Yang Y, Wang S, Yuan X, Yu H, Wang Y, Chen X, Liang X, Wei X (2018) Genome-wide association study of seed dormancy and the genomic consequences of improvement footprints in rice (Oryza sativa L.). Front Plant Sci 8:2213
Pritchard JK, Stephens M, Donnelly P (2000a) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000b) Association mapping in structured populations. Am J Hum Genet 67:170–181
Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Dai L, Han B, Wang G (2006) The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172:1901–1914
Riedelsheimer C, Lisec J, Czedik–Eysenberg A, Sulpice R, Flis A, Grieder C, Altmann T, Stitt M, Willmitzer L, Melchinger AE (2012) Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc Natl Acad Sci U S A 109:8872–8877
Rohlf FJ (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system, vol 21. Exeter Software, New York
Savary S, Horgan F, Willocquet L, Heong KL (2012) A review of principles for sustainable pest management in rice. Crop Prot 32:54–63
Shi LY, Li XH, Xie CX, Hao ZF, Weng JF, Zhang SH, Pan GT (2011) Development of SCARs from AFLP markers linked to resistance to maize rough dwarf virus (MRDV) using bulked segregant analysis in maize. Sci Agric Sin 9:1763–1774
Si L, Chen J, Huang X, Gong H, Luo J, Hou Q, Zhou T, Lu T, Zhu J, Shangguan Y, Chen E, Gong C, Zhao Q, Jing Y, Zhao Y, Li Y, Cui L, Fan D, Lu Y, Weng Q, Wang Y, Zhan Q, Liu K, Wei X, An K, An G, Han B (2016) OsSPL13 controls grain size in cultivated rice. Nat Genet 48:447–456
Sukumaran S, Dreisigacker S, Lopes M, Chavez P, Reynolds MP (2015) Genome-wide association study for grain yield and related traits in an elite spring wheat population grown in temperate irrigated environments. Theor Appl Genet 128:353–363
Tacconi G, Baldassarre V, Lanzanova C, Faivre-Rampant O, Cavigiolo S, Urso S, Lupotto E, Valè G (2010) Polymorphism analysis of genomic regions associated with broad-spectrum effective blast resistance genes for marker development in rice. Mol Breed 26:595–617
Takahashi A, Hayashi N, Miyao A, Hirochika H (2010) Unique features of the rice blast resistance Pish locus revealed by large scale retrotransposon-tagging. BMC Plant Biol 10:175
Tonnessen BW, Manosalva P, Lang JM, Baraoidan M, Bordeos A, Mauleon R, Oard J, Hulbert S, Leung H, Leach JE (2015) Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol Biol 87:273–286
Wang ZX, Yano M, Yamanouchi U, Iwamoto M, Monna L, Hayasaka H, Katayose Y, Sasaki T (1999) The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J 19:55–64
Wang C, Yang Y, Yuan X, Xu Q, Feng Y, Yu H, Wang Y, Wei X (2014) Genome-wide association study of blast resistance in indica rice. BMC Plant Biol 14:311
Wen Z, Tan R, Zhang S, Collins PJ, Yuan J, Du W, Gu C, Ou S, Song Q, An YC, Boyse JF, Chilvers M, Wang D (2018) Integrating GWAS and gene expression data for functional characterization of resistance to white mold in soybean. Plant Biotechnol J 16:1825–1836
Yu J, Pressoir G, Briggs WH, Vroh BI, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed–model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Zeng XS, Yang XF, Zhao ZH, Lin F, Wang L, Pan QH (2011) Characterization and fine mapping of the rice blast resistance gene Pia. Sci China Life Sci 54:372–378
Zhang Z, Ersoz E, Lai C, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, Buckler ES (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42:355–360
Zhang M, Lu Q, Wu W, Niu X, Wang C, Feng Y, Xu Q, Wang S, Yuan X, Yu H, Wang Y, Wei X (2017) Association mapping reveals novel genetic loci contributing to flooding tolerance during germination in indica rice. Front Plant Sci 8:678
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome 1:5–20
Funding
This research was financed by the Open Foundation of Guangdong Key Laboratory of Crops genetic improvement (2017B030314090 and 201801), the Special Foundation of President of the Guangdong Academy of Agricultural Sciences (201831), the National Natural Science Foundation of China (31801401), the Major Scientific and Technological Project for New Varieties Breeding of Zhejing Province (2016C02050-6-1), and the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-201X-CNRRI).
Author information
Authors and Affiliations
Contributions
XW designed the research; QL, XN, MZ, QX, YF, and SW performed the experiments; YH, HFL, and HL analyzed the data; QL wrote the manuscript; CW and YY edited the manuscript; XL and XW finalized the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Figure S1
(DOCX 420 kb)
Figure S2
(DOCX 2120 kb)
Figure S3
(DOCX 465 kb)
Figure S4
(DOCX 744 kb)
Figure S5
(DOCX 1834 kb)
Figure S6
(DOCX 138 kb)
Figure S7
(DOCX 210 kb)
Table S1
(XLSX 29 kb)
Table S2
(XLSX 11 kb)
Table S3
(XLSX 11 kb)
Table S4
(XLSX 26 kb)
Table S5
(XLSX 80 kb)
Table S6
(XLSX 19 kb)
Table S7
(XLSX 10 kb)
Table S8
(XLSX 10 kb)
Table S9
(XLSX 226 kb)
Table S10
(XLSX 10 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Lu, Q., Wang, C., Niu, X. et al. Detecting novel loci underlying rice blast resistance by integrating a genome-wide association study and RNA sequencing. Mol Breeding 39, 81 (2019). https://doi.org/10.1007/s11032-019-0989-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-0989-0