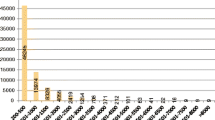
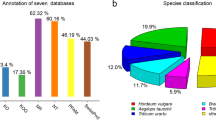
Abstract
Pennisetum purpureum belongs to the Pennisetum Rich genus in the family Poaceae. It is widely grown in subtropical and tropical regions as one of the most economically important cereal crops. Despite its importance, there is limited genomic data available for P. purpureum, which restricts genetic and breeding studies in this species. In the present study, the transcriptome of P. purpureum was assembled de novo and used to characterize two important P. purpureum cultivars: P. purpureum Schumab cv. Purple and P. purpureum cv. Mott. After assembly, a total of 197,466 unigenes were obtained for ‘Purple’ and ‘Mott’ and 103,454 of these unigenes were successfully annotated. From ‘Purple’ and ‘Mott,’ 214,648 SNPs and 21,213 EST-SSRs were identified in 40,259 unigenes and 18,587 unigenes respectively. Moreover, 50 EST-SSR primers and 6 SNP primers were designed to validate the identified markers. The transcriptomic data of present study from the two P. purpureum cultivars provides an abundant amount of available genomic information for Pennisetum. In addition, the identified SNPs and EST-SSRs will facilitate genetic and molecular studies within the Pennisetum genus.



Similar content being viewed by others
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Anderson WF, Dien BS, Brandon SK, Peterson JD (2008) Assessment of bermudagrass and bunch grasses as feedstock for conversion to ethanol. Appl Biochem Biotechnol 145:13–21
Babu C, Sundaramoorthi J, Vijayakumar G, Ram SG (2009) Analysis of genetic diversity in napier grass (Pennisetum purpureum Schum) as detected by RAPD and ISSR markers. J Plant Biochem Biotechnol 18:181–187
Barchi L, Lanteri S, Portis E, Acquadro A, Valè G, Toppino L, Rotino GL (2011) Identification of SNP and SSR markers in eggplant using RAD tag sequencing. BMC Genomics 12:304
Bhandari AP, Sukanya D, Ramesh C (2006) Application of isozyme data in fingerprinting Napier grass (Pennisetum purpureum Schum.) for germplasm management. Genet Resour Crop Evol 53:253–264
Bragg JG, Supple MA, Andrew RL, Borevitz JO (2015) Genomic variation across landscapes: insights and applications. New Phytol 207:953–967
Brunken JN (1977) A systematic study of Pennisetum sect. Pennisetum (Gramineae). Am J Bot:161–176
Campos J et al (2009) In vitro induction of hexaploid plants from triploid hybrids of Pennisetum purpureum and Pennisetum glaucum. Plant Breed 128:101–104
Choi JY, Bubnell JE, Aquadro CF (2015) Population genomics of infectious and integrated Wolbachia pipientis genomes in Drosophila ananassae. Genome Biol Evol 7:2362–2382
Choudhary M, Padaria JC (2015) Transcriptional profiling in pearl millet (Pennisetum glaucum LR Br.) for identification of differentially expressed drought responsive genes. Physiol Mol Biol Plants 21:187–196
Coulondre C, Miller JH, Farabaugh PJ, Gilbert W (1978) Molecular basis of base substitution hotspots in Escherichia coli. Nature 274:775
Du F et al (2015) De novo assembled transcriptome analysis and SSR marker development of a mixture of six tissues from Lilium Oriental hybrid ‘Sorbonne’. Plant Mol Biol Report 33:281–293
Dutta S et al (2011) Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh]. BMC Plant Biol 11(17)
Eckert AJ, Pande B, Ersoz ES, Wright MH, Rashbrook VK, Nicolet CM, Neale DB (2009) High-throughput genotyping and mapping of single nucleotide polymorphisms in loblolly pine (Pinus taeda L.). Tree Genet Genomes 5:225–234
Egan AN, Schlueter J, Spooner DM (2012) Applications of next-generation sequencing in plant biology. Am J Bot 99:175–185
Ekblom R, Galindo J (2011) Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 107(1)
Fulkerson W, Horadagoda A, Neal J, Barchia I, Nandra K (2008) Nutritive value of forage species grown in the warm temperate climate of Australia for dairy cows: herbs and grain crops. Livest Sci 114:75–83
Gaur R et al (2012) High-throughput SNP discovery and genotyping for constructing a saturated linkage map of chickpea (Cicer arietinum L.). DNA Res 19:357–373
Gilles A, Meglécz E, Pech N, Ferreira S, Malausa T, Martin J-F (2011) Accuracy and quality assessment of 454 GS-FLX titanium pyrosequencing. BMC Genomics 12:245
Grabherr MG et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644
Guo Z-H et al (2016) SSRs transferability and genetic diversity of three allogamous ryegrass species. C R Biol 339:60–67
Gupta D et al (2012) Integration of EST-SSR markers of Medicago truncatula into intraspecific linkage map of lentil and identification of QTL conferring resistance to ascochyta blight at seedling and pod stages. Mol Breed 30:429–439
Ha B-K, Hussey RS, Boerma HR (2007) Development of SNP assays for marker-assisted selection of two southern root-knot nematode resistance QTL in soybean. Crop Sci 47:S-73–S-82
Harris K, Anderson W, Malik R (2010) Genetic relationships among napiergrass (Pennisetum purpureum Schum.) nursery accessions using AFLP markers. Plant Genet Resour 8:63–70
Hayashi K, Hashimoto N, Daigen M, Ashikawa I (2004) Development of PCR-based SNP markers for rice blast resistance genes at the Piz locus. Theor Appl Genet 108:1212–1220
Hua W, Zheng P, He Y, Cui L, Kong W, Wang Z (2014) An insight into the genes involved in secoiridoid biosynthesis in Gentiana macrophylla by RNA-seq. Mol Biol Rep 41:4817–4825
Huang X et al (2014) Genetic diversity of Hemarthria altissima and its related species by EST-SSR and SCoT markers. Biochem Syst Ecol 57:338–344
Huang L et al (2015) Identifying differentially expressed genes under heat stress and developing molecular markers in orchardgrass (Dactylis glomerata L.) through transcriptome analysis. Mol Ecol Resour 15:1497–1509
Huang J, Gao Y, Jia H, Zhang Z (2016a) Characterization of the teosinte transcriptome reveals adaptive sequence divergence during maize domestication. Mol Ecol Resour 16:1465–1477
Huang J et al (2016b) De novo sequencing and characterization of seed transcriptome of the tree legume Millettia pinnata for gene discovery and SSR marker development. Mol Breed 36(75)
Huang X et al (2016c) De novo transcriptome analysis and molecular marker development of two Hemarthria species. Front Plant Sci 7:496
Iseli C, Jongeneel CV, Bucher P (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. In: ISMB, pp 138–148
Jakob K, Zhou F, Paterson AH (2009) Genetic improvement of C4 grasses as cellulosic biofuel feedstocks. In Vitro Cell Dev Biol: Plant 45:291–305
Jhanwar S, Priya P, Garg R, Parida SK, Tyagi AK, Jain M (2012) Transcriptome sequencing of wild chickpea as a rich resource for marker development. Plant Biotechnol J 10:690–702
Jia X, Deng Y, Sun X, Liang L, Su J (2016) De novo assembly of the transcriptome of Neottopteris nidus using Illumina paired-end sequencing and development of EST-SSR markers. Mol Breed 36:94
Jiang Q, Wang F, Tan H-W, Li M-Y, Xu Z-S, Tan G-F, Xiong A-S (2015) De novo transcriptome assembly, gene annotation, marker development, and miRNA potential target genes validation under abiotic stresses in Oenanthe javanica. Mol Gen Genomics 290:671–683
Jiang F, Chen X-p, Hu W-s, Zheng S-q (2016) Identification of differentially expressed genes implicated in peel color (red and green) of Dimocarpusconfinis. SpringerPlus 5:1088
Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48:501–510
Kaur S et al (2012) Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers. BMC Genomics 13:104
Kawube G, Alicai T, Wanjala B, Njahira M, Awalla J, Skilton R (2015) Genetic diversity in Napier grass (Pennisetum purpureum) assessed by SSR markers. J Agric Sci 7:147
Khairwal I, Rai K, Diwakar B, Sharma Y, Rajpurohit B, Nirwan B, Bhattacharjee R (2007) Pearl millet crop management and seed production manual. International Crops Research Institute for the Semi-Arid Tropics, Patancheru
Lakew B, Henry RJ, Ceccarelli S, Grando S, Eglinton J, Baum M (2013) Genetic analysis and phenotypic associations for drought tolerance in Hordeum spontaneum introgression lines using SSR and SNP markers. Euphytica 189:9–29
Lee M-K, Tsai W-T, Tsai Y-L, Lin S-H (2010) Pyrolysis of napier grass in an induction-heating reactor. J Anal Appl Pyrolysis 88:110–116
Li Y-C, Korol AB, Fahima T, Nevo E (2004) Microsatellites within genes: structure, function, and evolution. Mol Biol Evol 21:991–1007
Li H et al (2009a) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li Y-H et al (2009b) Development of SNP markers and haplotype analysis of the candidate gene for rhg1, which confers resistance to soybean cyst nematode in soybean. Mol Breed 24:63–76
Li H, Yao W, Fu Y, Li S, Guo Q (2015) De novo assembly and discovery of genes that are involved in drought tolerance in Tibetan Sophora moorcroftiana. PLoS One 10:e111054
Liu B, Zhang Y, Zhang W (2014) RNA-Seq-based analysis of cold shock response in Thermoanaerobacter tengcongensis, a bacterium harboring a single cold shock protein encoding gene. PLoS One 9:e93289
Lowe A, Thorpe W, Teale A, Hanson J (2003) Characterisation of germplasm accessions of Napier grass (Pennisetum purpureum and P. purpureum× P. glaucum hybrids) and comparison with farm clones using RAPD. Genet Resour Crop Evol 50:121–132
Lv J, Liu P, Gao B, Wang Y, Wang Z, Chen P, Li J (2014) Transcriptome analysis of the Portunus trituberculatus: de novo assembly, growth-related gene identification and marker discovery. PLoS One 9:e94055
McCallum S et al (2016) Construction of a SNP and SSR linkage map in autotetraploid blueberry using genotyping by sequencing. Mol Breed 36(41)
Metzker ML (2010) Sequencing technologies—the next generation. Nat Rev Genet 11(31)
Morais RF, Souza BJ, Leite JM, Soares LHB, Alves BJR, Boddey RM, Urquiaga S (2009) Elephant grass genotypes for bioenergy production by direct biomass combustion. Pesq Agrop Brasileira 44:133–140
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185
Moumouni K, Kountche B, Jean M, Hash C, Vigouroux Y, Haussmann B, Belzile F (2015) Construction of a genetic map for pearl millet, Pennisetum glaucum (L.) R. Br., using a genotyping-by-sequencing (GBS) approach. Mol Breed 35(5)
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci 70:3321–3323
Ockendon NF et al (2016) Optimization of next-generation sequencing transcriptome annotation for species lacking sequenced genomes. Mol Ecol Resour 16:446–458
Pavy N, Parsons LS, Paule C, MacKay J, Bousquet J (2006) Automated SNP detection from a large collection of white spruce expressed sequences: contributing factors and approaches for the categorization of SNPs. BMC Genomics 7:174
Pei M, Niu J, Li C, Cao F, Quan S (2016) Identification and expression analysis of genes related to calyx persistence in Korla fragrant pear. BMC Genomics 17:132
Punta M et al (2011) The Pfam protein families database. Nucleic Acids Res 40:D290–D301
Qiao Q et al (2016) Comparative transcriptomics of strawberries (Fragaria spp.) provides insights into evolutionary patterns. Front Plant Sci 7:1839
Rafalski A (2002) Applications of single nucleotide polymorphisms in crop genetics. Curr Opin Plant Biol 5:94–100
Rai K, Gupta S, Ranjana B, Kulkarni V, Singh A, Rao A (2009) Morphological characteristics of ICRISAT-bred pearl millet hybrid seed parents. Andhra Pradesh, India, pp 4
Rajaram V et al (2013) Pearl millet [Pennisetum glaucum (L.) R. Br.] consensus linkage map constructed using four RIL mapping populations and newly developed EST-SSRs. BMC Genomics 14:159
Riahi L et al (2013) Characterization of single nucleotide polymorphism in Tunisian grapevine genome and their potential for population genetics and evolutionary studies. Genet Resour Crop Evol 60:1139–1151
Sahu PP, Gupta S, Malaviya D, Roy AK, Kaushal P, Prasad M (2012) Transcriptome analysis of differentially expressed genes during embryo sac development in apomeiotic non-parthenogenetic interspecific hybrid of Pennisetum glaucum. Mol Biotechnol 51:262–271
Salem M et al (2012) RNA-Seq identifies SNP markers for growth traits in rainbow trout. PLoS One 7:e36264
Sarah G et al (2017) A large set of 26 new reference transcriptomes dedicated to comparative population genomics in crops and wild relatives. Mol Ecol Resour 17:565–580
Schmid KJ, Sörensen TR, Stracke R, Törjék O, Altmann T, Mitchell-Olds T, Weisshaar B (2003) Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res 13:1250–1257
Singh RK et al (2013) Development, cross-species/genera transferability of novel EST-SSR markers and their utility in revealing population structure and genetic diversity in sugarcane. Gene 524:309–329
Soren KR et al (2015) EST-SSR analysis provides insights about genetic relatedness, population structure and gene flow in grass pea (Lathyrus sativus). Plant Breed 134:338–344
Sousa Azevedo AL, Costa PP, Machado JC, Machado MA, Pereira AV, José da Silva Lédo F (2012) Cross species amplification of Pennisetum glaucum microsatellite markers in Pennisetum purpureum and genetic diversity of Napier grass accessions. Crop Sci 52:1776–1785
Strezov V, Evans TJ, Hayman C (2008) Thermal conversion of elephant grass (Pennisetum Purpureum Schum) to bio-gas, bio-oil and charcoal. Bioresour Technol 99:8394–8399
Sujatha D, Manga V, Rao M, Murty J (1989) Meiotic studies in some species of Pennisetum (L.) rich. (Poaceae). Cytologia 54:641–652
Sureshkumar S et al (2014) Marker-assisted introgression of lpa2 locus responsible for low-phytic acid trait into an elite tropical maize inbred (Zea mays L.). Plant Breed 133:566–578
Takayama K, López PS, König C, Kohl G, Novak J, Stuessy TF (2011) A simple and cost-effective approach for microsatellite isolation in non-model plant species using small-scale 454 pyrosequencing. Taxon 60:1442–1449
Tang X, Xiao Y, Lv T, Wang F, Zhu Q, Zheng T, Yang J (2014) High-throughput sequencing and de novo assembly of the Isatis indigotica transcriptome. PLoS One 9:e102963
Taylor M, Vasil I (1987) Analysis of DNA size, content and cell cycle in leaves of Napier grass (Pennisetum purpureum Schum.). Theor Appl Genet 74:681–686
Toledo-Silva G, Cardoso-Silva CB, Jank L, Souza AP (2013) De novo transcriptome assembly for the tropical grass Panicum maximum Jacq. PLoS One 8:e70781
Ukoskit K, Posudsavang G, Pongsiripat N, Chatwachirawong P, Klomsa-ard P, Poomipant P, Tragoonrung S (2018) Detection and validation of EST-SSR markers associated with sugar-related traits in sugarcane using linkage and association mapping. Genomics. https://doi.org/10.1016/j.ygeno.2018.03.019
Varshney R et al (2008) Identification and validation of a core set of informative genic SSR and SNP markers for assaying functional diversity in barley. Mol Breed 22:1–13
Wang M, Yan J, Zhao J, Song W, Zhang X, Xiao Y, Zheng Y (2012) Genome-wide association study (GWAS) of resistance to head smut in maize. Plant Sci 196:125–131
Wang X, Li S, Li J, Li C, Zhang Y (2015) De novo transcriptome sequencing in Pueraria lobata to identify putative genes involved in isoflavones biosynthesis. Plant Cell Rep 34:733–743
Wang Y et al (2017) Development of SNP markers based on transcriptome sequences and their application in germplasm identification in radish (Raphanus sativus L.). Mol Breed 37(26)
Wanjala BW et al (2013) Genetic diversity in Napier grass (Pennisetum purpureum) cultivars: implications for breeding and conservation. AoB Plants 5
Wei L et al (2014) Development of SNP and InDel markers via de novo transcriptome assembly in Sesamum indicum L. Mol Breed 34:2205–2217
Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Wu J et al (2014a) High-density genetic linkage map construction and identification of fruit-related QTLs in pear using SNP and SSR markers. J Exp Bot 65:5771–5781
Wu Z-J, Li X-H, Liu Z-W, Xu Z-S, Zhuang J (2014b) De novo assembly and transcriptome characterization: novel insights into catechins biosynthesis in Camellia sinensis. BMC Plant Biol 14:277
Xie X-M, Lu X-L (2005) Analysis of genetic relationships of cultivars in Pennisetum by RAPD markers. Acta Pratacultural Science 2
Xie X-M, Zhou F, Zhang X-Q, Zhang J-M (2009) Genetic variability and relationship between MT-1 elephant grass and closely related cultivars assessed by SRAP markers. J Genet 88:281–290
Xu P et al (2011) A SNP and SSR based genetic map of asparagus bean (Vigna. unguiculata ssp. sesquipedialis) and comparison with the broader species. PLoS One 6:e15952
Xu M, Liu X, Wang J-W, Teng S-Y, Shi J-Q, Li Y-Y, Huang M-R (2017) Transcriptome sequencing and development of novel genic SSR markers for Dendrobium officinale. Mol Breed 37(18)
Yang X, Hang X, Tan J, Yang H (2015) Differences in acid tolerance between Bifidobacterium breve BB8 and its acid-resistant derivative B. breve BB8dpH, revealed by RNA-sequencing and physiological analysis. Anaerobe 33:76–84
Yao YF, Hong JJ, Zeng RQ (2013) SRAP analysis on genetic diversity of Pennisetum. J Gansu Agric Univ 4:108–109
Yates SA et al (2014) De novo assembly of red clover transcriptome based on RNA-Seq data provides insight into drought response, gene discovery and marker identification. BMC Genomics 15:453
Yeh F (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot 129:157
Yue X, Nie Q, Xiao G, Liu B (2015) Transcriptome analysis of shell color-related genes in the clam Meretrix meretrix. Mar Biotechnol 17:364–374
Zeng A et al (2017) Genome-wide association study (GWAS) of salt tolerance in worldwide soybean germplasm lines. Mol Breed 37(30)
Zhang Y, Cheng Y, Ya H, Han J, Zheng L (2015a) Identification of heat shock proteins via transcriptome profiling of tree peony leaf exposed to high temperature. Genet Mol Res 14:8431–8442
Zhang Z, Pang T, Li Q, Zhang L, Li L, Liu J (2015b) Transcriptome sequencing and characterization for Kappaphycus alvarezii. Eur J Phycol 50:400–407
Zhang W, Guo Y, Li J, Huang L, Kazitsa EG, Wu H (2016a) Transcriptome analysis reveals the genetic basis underlying the seasonal development of keratinized nuptial spines in Leptobrachium boringii. BMC Genomics 17:978
Zhang Y, Tao S, Yuan C, Liu Y, Wang Z (2016b) Non-monotonic dose–response effect of bisphenol A on rare minnow Gobiocypris rarus ovarian development. Chemosphere 144:304–311
Zhao W et al (2014) RNA-Seq-based transcriptome profiling of early nitrogen deficiency response in cucumber seedlings provides new insight into the putative nitrogen regulatory network. Plant Cell Physiol 56:455–467
Zhou Z et al (2014) Transcriptome sequencing of sea cucumber (Apostichopus japonicus) and the identification of gene-associated markers. Mol Ecol Resour 14:127–138
Zou D, Chen X, Zou D (2013) Sequencing, de novo assembly, annotation and SSR and SNP detection of sabaigrass (Eulaliopsis binata) transcriptome. Genomics 102:57–62
Acknowledgements
We thank Tomas Hasing Rodriguez who helped polish this manuscript.
Funding
The authors gratefully acknowledge the financial support from the National Project on Sci-Tec Foundation Resources Survey (2017FY100602), Sichuan Province Breeding Research Grant (2016NZ0098-11), and Modern Agricultural Industry System Sichuan Forage Innovation Team.
Author information
Authors and Affiliations
Contributions
L.K.H. and X.Q.Z. designed research studies; S.F.Z., C.R.W., and H.D.Y. conducted experiments, acquired data, and analyzed data; L.K.H., X.Q.Z., Y.P., X.M., and Y.H.Y. provided the experimental equipment; S.F.Z. and P.L.C. wrote the manuscript; T.P.F. and Z.H.C. revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Data archiving statement
Raw Illumina reads were deposited in NCBI SRA: SRP100008.
Electronic supplementary material
Fig. S1
The length distribution of CDSs mapped to known genes by BLAST. (PNG 89 kb)
Fig. S2
The length distribution of CDSs mapped to known genes by ESTScan. (PNG 85 kb)
Figure S3
UPGMA dendrogram based on Nei’s genetic diversity between the seventeen Pennisetum species. (PNG 258 kb)
Table S1
(DOCX 74 kb)
Table S2
(DOCX 64 kb)
Table S3
(DOCX 23 kb)
Table S4
(DOCX 53 kb)
Table S5
(DOCX 36 kb)
Table S6
(DOCX 36 kb)
Table S7
(DOCX 40 kb)
Table S8
(DOCX 65 kb)
Table S9
(DOCX 47 kb)
Table S10
(DOCX 33 kb)
Table S11
(DOCX 84 kb)
Rights and permissions
About this article
Cite this article
Zhou, S., Wang, C., Frazier, T.P. et al. The first Illumina-based de novo transcriptome analysis and molecular marker development in Napier grass (Pennisetum purpureum). Mol Breeding 38, 95 (2018). https://doi.org/10.1007/s11032-018-0852-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-018-0852-8