Abstract
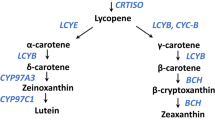
The common flesh color of commercially grown watermelon is red due to the accumulation of lycopene. However, natural variation in carotenoid composition that exists among heirloom and exotic accessions results in a wide spectrum of flesh colors. We previously identified a unique orange flesh watermelon accession (NY0016) that accumulates mainly β-carotene and no lycopene. We hypothesized this unique accession could serve as a viable source for increasing provitamin A content in watermelon. Here we characterize the mode of inheritance and genetic architecture of this trait. Analysis of testcrosses of NY0016 with yellow and red fruited lines indicated a codominant mode of action as F1 fruits exhibited a combination of carotenoid profiles from both parents. We combined visual color phenotyping with genotyping-by-sequencing of an F2:3 population from a cross of NY0016 by a yellow fruited line, to map a major locus on chromosome 1, associated with β-carotene accumulation in watermelon fruit. The QTL interval is approximately 20 cM on the genetic map and 2.4 Mb on the watermelon genome. Trait-linked marker was developed and used for validation of the QTL effect in segregating populations across different genetic backgrounds. This study is a step toward identification of a major gene involved in carotenoid biosynthesis and accumulation in watermelon. The codominant inheritance of β-carotene provides opportunities to develop, through marker-assisted breeding, β-carotene-enriched red watermelon hybrids.





Similar content being viewed by others
References
Bang H, Davis AR, Kim S et al (2010) Flesh color inheritance and gene interactions among canary yellow, pale yellow, and red watermelon. J Am Soc Hortic Sci 135(4):362–368
Bang H, Kim S, Leskovar D, King S (2007) Development of a codominant CAPS marker for allelic selection between canary yellow and red watermelon based on SNP in lycopene β-cyclase (LCYB) gene. Mol Breed 20:63–72
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57:289–300
Bradbury PJ, Zhang Z, Kroon DE et al (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Bramley PM (2000) Is lycopene beneficial to human health? Phytochemistry 54:233–236
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Broman KW, Sen S (2009) A guide to QTL mapping with R/qtl. Vol. 46. Springer, New York
Carlborg O, Haley CS (2004) Epistasis: too often neglected in complex trait studies? Nat Rev Genet 5:618–625
Elshire RJ, Glaubitz JC, Sun Q et al (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379
Gerster H (1997) The potential role of lycopene for human health. J Am Coll Nutr 16:109–126
Giovannucci E, Rimm EB, Liu Y, Stampfer MJ, Willett WC (2002) A prospective study of tomato products, lycopene, and prostate cancer risk. J Natl Cancer Inst 94:391–398
Glaubitz JC, Casstevens TM, Lu F et al (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9:e90346
Guo S, Sun H, Zhang H et al (2015) Comparative transcriptome analysis of cultivated and wild watermelon during fruit development. PLoS One 10:1–21
Guo S, Zhang J, Sun H et al (2013) The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet 45:51–58
Gusmini G, Wehner TC (2006) Qualitative inheritance of rind pattern and flesh color in watermelon. J Hered 97:177–185
Haley CS, Knott SA (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69:315–324
Hashizume T, Shimamoto I, Hirai M (2003) Construction of a linkage map and QTL analysis of horticultural traits for watermelon [Citrullus lanatus (THUNB.) MATSUM & NAKAI] using RAPD, RFLP and ISSR markers. Theor Appl Genet 106:779–785
Henderson WR (1989) Inheritance of orange flesh color in watermelon. Cucurbit Genet Coop Rpt 12:59–63
Henderson WR, Scott GH, Wehner TC (1998) Interaction of flesh color genes in watermelon. J Hered 89:50–53
Kim OR, Cho MC, Kim BD, Huh JH (2010) A splicing mutation in the gene encoding phytoene synthase causes orange coloration in habanero pepper fruits. Molecules and Cells 30:569–574
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugenics 12:172–175
Kruglyak L, Lander ES (1995) A nonparametric approach for mapping quantitative trait loci. Genetics 139:1421–1428
Kruskal WH, Wallis WA (1952) Use of ranks in one-criterion variance analysis. J Am Stat Assoc 47(260):583–621
Lambel S, Lanini B, Vivoda E et al (2014) A major QTL associated with Fusarium oxysporum race 1 resistance identified in genetic populations derived from closely related watermelon lines using selective genotyping and genotyping-by-sequencing for SNP discovery. Theor Appl Genet 127(10):2105–2115
Lander ES, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci U S A 84:2363–2367
Lewinsohn E, Sitrit Y, Bar E et al (2005a) Not just colors—carotenoid degradation as a link between pigmentation and aroma in tomato and watermelon fruit. Trends Food Sci Technol 16:407–415
Lewinsohn E, Sitrit Y, Bar E et al (2005b) Carotenoid pigmentation affects the volatile composition of tomato and watermelon fruits, as revealed by comparative genetic analyses. J Agric Food Chem 53:3142–3148
Liu S, Gao P, Wang X et al (2015) Mapping of quantitative trait loci for lycopene content and fruit traits in Citrullus lanatus. Euphytica 202:411–426
Liu S, Gao P, Zhu Q et al (2016) Development of cleaved amplified polymorphic sequence markers and a CAPS-based genetic linkage map in watermelon (Citrullus lanatus [Thunb.] Matsum. and Nakai) constructed using whole-genome re-sequencing data. Breed Sci 66:244–259
Lv P, Li N, Liu H et al (2015) Changes in carotenoid profiles and in the expression pattern of the genes in carotenoid metabolisms during fruit development and ripening in four watermelon cultivars. Food Chem 174:52–59
Manichaikul A, Moon JY, Sen Ś et al (2009) A model selection approach for the identification of quantitative trait loci in experimental crosses, allowing epistasis. Genetics 181:1077–1086
Meru G, McGregor C (2016) Genotyping by sequencing for SNP discovery and genetic mapping of resistance to race 1 of Fusarium oxysporum in watermelon. Sci Hortic 209:31–40
Perkins-Veazie P, Collins JK, Davis AR, Roberts W (2006) Carotenoid content of 50 watermelon cultivars. J Agric Food Chem 54:2593–2597
Poole CF (1944) Genetics of cultivated cucurbits. J Hered 35(4):122–128
Porter DR (1937) Inheritance of certain fruit and seed characters in watermelons. Hilgardia 10(12):489–509
Development Core Team R (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Reddy UK, Nimmakayala P, Levi A, et al (2014) High-resolution genetic map for understanding the effect of genome-wide recombination rate on nucleotide diversity in watermelon. G3 (Bethesda, Md) 4:2219–30
Ronen G, Cohen M, Zamir D, Hirschberg J (1999) Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J 17:341–351
Ronen G, Carmel-Goren L, Zamir D, Hirschberg J (2000) An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. PNAS 97:11102–11107
Sandlin K, Prothro J, Heesacker A et al (2012) Comparative mapping in watermelon [Citrullus lanatus (Thunb.) Matsum. et Nakai]. Theor Appl Genet 125:1603–1618
Shang J, Li N, Li N et al (2016) Construction of a high-density genetic map for watermelon (Citrullus lanatus L.) based on large-scale SNP discovery by specific length amplified fragment sequencing (SLAF-seq). Sci Hortic 203:38–46
Shimotsuma M (1963) Cytogenetical studies in the genus Citrullus: VII. Inheritance of several characters in watermelons. Jpn J Breed 13(4):235–240
Swarts K, Li H, Romero Navarro JA et al (2014) Novel methods to optimize genotypic imputation for low-coverage, next-generation sequence data in crop plants. Plant Genome 7. https://doi.org/10.3835/plantgenome2014.05.0023
Tadmor Y, Katzir N, King S, Levi A, Davis A, Hirschberg J (2004) Fruit coloration in watermelon: lessons from the tomato. In: Lebeda A, Paris HS (eds) Progress in cucurbit genetics and breeding research. Palacky University in Olomouc, Czech Republic, pp 181–185
Tadmor Y, King S, Levi A et al (2005) Comparative fruit colouration in watermelon and tomato. Food Res Int 38:837–841
Wang N, Liu S, Gao P et al (2016) Developmental changes in gene expression drive accumulation of lycopene and β-carotene in watermelon. J Am Soc Hortic Sci 141:434–443
Wehner TC (2007) Gene list for watermelon. Cucurbit Genet Coop Rep 30:96–120
Welsch R, Arango J, Bär C, Salazar B, Al-Babili S, Beltrán J, Chavarriaga P, Ceballos H, Tohme J, Beyer P (2010) Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell 22:3348–3356
Zeng ZB, Kao CH, Basten CJ (1999) Estimating the genetic architecture of quantitative traits. Genet Res 74:279–289
Acknowledgements
We gratefully acknowledge support from the “Center for the Improvement of Cucurbit Fruit Quality,” ARO, Israel. We also acknowledge partial support by US-Israel Binational Agricultural Research & Development (BARD) Senior Research Fellowship Program (USDA-BARD Agreement Number 58-6080-5-011F), and by USDA-National Institute of Food and Agriculture (NIFA), Specialty Crop Research Initiative (SCRI) grant number 2015-51181-24285. We also thank the USA National Watermelon Research and Promotion Board for their continual support; publication no. 211/2017 of the Agricultural Research Organization, Bet Dagan, Israel.
Funding
Funding for this research was provided by the Israeli Ministry of Agriculture Chief Scientist grant no. 20-01-0135.
Author information
Authors and Affiliations
Contributions
AG, AL, AM, and YT conceived and designed the study. LV, AM, and GT performed field experiments, phenotyping, and genotyping. AG, ZF, and SB analyzed the data. AG, SB, AL, WW, and YT wrote the paper. All authors discussed the results and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Key message
Major QTL regulating the accumulation of β-carotene in the fruit flesh was mapped to 2 Mbp intervals on chromosome 1 in watermelon.
Electronic supplementary material
ESM 1
Figure S1. Photographs of longitudinal cross-sections of representative individuals for each flesh color category and a pie chart of segregation ratios for the (a) F2 population and (b) F2:3 families. Figures S2-S3. Genetic linkage map of Citrullus lanatus with 1078 binned SNPs produced from an F2:3 population derived from experimental line NY0016 crossed with cultivar EMB. Linkage groups 1 through 6 are displayed in Figure S1 and linkage groups 7–11 in Figure S2. SNP positions (cM) are labelled to the left of each linkage group (labelled at the top of the figure) and the chromosome and physical position (bp) are to the right. The 1.5-LOD interval of qFlesh-1 is indicated with a pink vertical bar to the right of linkage group 1. Fig. S4. Plots of genetic (cM) versus physical distance (bp) of each SNP across the 11 watermelon chromosomes. Fig. S5. Gel electrophoresis of PCR with CAPS marker for the parents (EMB and NY0016), F1, and representative F2 individuals. Letters above the gel bands represent the translated genotype: E = Homozygote EMB, N = Homozygote NY0016, H = Heterozygote. (PDF 1735 kb)
Table S1
Variation in color and carotenoid concentrations in four testcrosses with NY0016 (XLSX 10 kb)
Table S2
Flesh color data for the F2:3 families in categorical and continuous scales (separated tabs), including pedigree, family ID (family); the family flesh color categories used for QTL mapping (flesh0–1), family categorical flesh ratings (seg) and the individual fruit colors (fruit1-fruit10). (XLSX 23 kb)
Table S3
1078 Binned SNPs information, including: SNP name (SNP), genetic position (cM) on each linkage group (LG), physical position (bp) on each chromosome (CS), results of a chi test for segregation distortion (P, neglog10P, and FDR), and the proportions of individuals with each genotype at that locus (AA, AB, BB, or missing). AA indicates individuals homozygous for NY0016 alleles, BB homozygous for EMB alleles or heterozygous individuals AB. (XLSX 111 kb)
Table S4
Annotated genes list within the 1.5-LOD interval (8.00 to 10.50 Mb on chromosome 1) for flesh color QTL (qFC.1). Gene information includes: identifier for watermelon gene (geneID), chromosome (CG_Chr), start position (bp), stop position (bp), and the functional annotation. Annotated candidates are highlighted. (XLSX 18 kb)
Rights and permissions
About this article
Cite this article
Branham, S., Vexler, L., Meir, A. et al. Genetic mapping of a major codominant QTL associated with β-carotene accumulation in watermelon. Mol Breeding 37, 146 (2017). https://doi.org/10.1007/s11032-017-0747-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-017-0747-0