Abstract
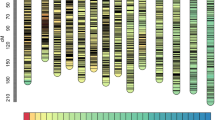
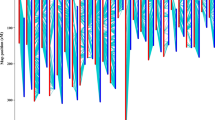
We used RAD (restriction-site-associated DNA) sequencing to detect genome-wide SNPs and construct a dense linkage map using an intercross F2 population in jute (Corchorus olitorius). The linkage map comprising a total of 503 RAD markers in seven linkage groups spanned 358.5 cM with an average marker interval of 0.72 cM and covered 87.0 % of the genome. Genome-wide segregation distortion of the mapped loci (34.4 %) was non-random across the linkage map, with a directional bias mostly towards the female genotypes. Jute had maximum syntenic relationships with cocoa (47.5 % homology) and diploid cotton (29.2 % homology). However, synteny and collinearity were not conserved. Histological fibre content (FC; total number of fibre cell bundles in a stem cross section) was positively correlated with fibre yield (FY), plant height (PH), root weight (RW) and stem-base diameter (SBD). Broad-sense heritability estimates were high for all traits, with FC and FY showing maximum heritability (~93 %). QTL mapping based on the F2:3 phenotypes detected nine QTL across the two environments. The QTL for FC was coincident with one QTL each for FY, PH, RW and SBD on top of a single-SNP (C/T) marker at 40.2 cM on LG1, each accounting for ~7–11 % of the phenotypic variance. Two QTL linked in repulsion one each for PH and SBD, with varying degrees of overdominance, were associated with two single-SNP (C/T) markers on LG2, each accounting for ~17–18 % of the phenotypic variance. Few candidate genes were identified within the QTL regions. Our results would enable development of tools for marker-assisted selection in jute.



Similar content being viewed by others
Abbreviations
- bfs:
-
Bast fibre-shy
- FC:
-
Fibre content
- FCB:
-
Fibre cell bundle
- FY:
-
Fibre yield
- LG:
-
Linkage group
- PH:
-
Plant height
- QTL:
-
Quantitative trait loci
- RAD-seq:
-
Restriction-site-associated DNA sequencing
- RW:
-
Root weight
- SBD:
-
Stem-base diameter
- SG:
-
Sudan Green
- SNP:
-
Single nucleotide polymorphism
References
Alverson WS, Whitlock BA, Nyffeler R, Bayer C, Baum DA (1999) Phylogeny of the core Malvales: evidence from ndhF sequence data. Am J Bot 86:1474–1486
Argout X, Salse J, Aury J-M, Guiltinan MJ, Droc G, Gouzy J, Allegre M, Chaparro C et al (2011) The genome of Theobroma cacao. Nat Genet 43:101–108. doi:10.1038/ng.736
Arunachalam V, Iyer RD (1978) A non-destructive selection criterion for fibre content in jute. III. The criterion and its prospects. Theor Appl Genet 52:129–134. doi:10.1007/bf00264746
Bai C, Liang Y, Hawkesford MJ (2013) Identification of QTLs associated with seedling root traits and their correlation with plant height in wheat. J Exp Bot 64:1745–1753. doi:10.1093/jxb/ert041
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3:e3376. doi:10.1371/journal.pone.0003376
Barchi L, Lanteri S, Portis E, Acquadro A, Valè G, Toppino L, Rotino GL (2011) Identification of SNP and SSR markers in eggplant using RAD tag sequencing. BMC Genom 12:304. doi:10.1186/1471-2164-12-304
Barchi L, Lanteri S, Portis E, Valè G, Volante A, Pulcini L, Ciriaci T, Acciarri N, Barbierato V, Toppino L, Rotino GL (2012) A RAD tag derived marker based eggplant linkage map and the location of QTLs determining anthocyanin pigmentation. PLoS One 7:e43740. doi:10.1371/journal.pone.0043740
Basak SL (1993) Quantitative genetics of fibre yield and its components. In: Denton IR (ed) Review on the genetics and breeding of jute. International Jute Organization, Dhaka, pp 51–95
Begum R, Zakrzewski F, Menzel G, Weber B, Alam SS, Schmidt T (2013) Comparative molecular cytogenetic analyses of a major tandemly repeated DNA family and retrotransposon sequences in cultivated jute Corchorus species (Malvaceae). Ann Bot 112:123–134. doi:10.1093/aob/mct103
Benor S, Fuchs J, Blattner FR (2011) Genome size variation in Corchorus olitorius (Malvaceae s. l.) and its correlation with elevation and phenotypic traits. Genome 54:575–585. doi:10.1139/g11-021
Benor S, Demissew S, Hammer K, Blattner FR (2012) Genetic diversity and relationships in Corchorus olitorius (Malvaceae s. l.) inferred from molecular and morphological data. Genet Resour Crop Evol 59:1125–1146. doi:10.1007/s10722-011-9748-8
Bus A, Hecht J, Huettel B, Reinhardt R, Stich B (2012) High-throughput polymorphism detection and genotyping in Brassica napus using next-generation RAD sequencing. BMC Genom 13:281. doi:10.1186/1471-2164-13-281
Capron A, Chang XF, Hall H, Ellis B, Beatson RP, Berleth T (2013) Identification of quantitative trait loci controlling fibre length and lignin content in Arabidopsis thaliana stems. J Exp Bot 64:185–197. doi:10.1093/jxb/ers319
Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH (2011) Stacks: building and genotyping loci de novo from short-read sequences. G3 1:171–182. doi:10.1534/g3.111.000240
Chakravarti A, Lasher LK, Reefer JE (1991) A maximum likelihood method for estimating genome length using genetic linkage data. Genetics 128:175–182
Chen MX, Wei CH, Qi JM, Chen XB, Su JG, Li AQ, Tao AF, Wu WR (2011) Genetic linkage map construction for kenaf using SRAP, ISSR and RAPD markers. Plant Breed 130:679–687. doi:10.1111/j.1439-0523.2011.01879.x
Chutimanitsakun Y, Nipper RW, Cuesta-Marcos A, Cistué L, Corey A, Filichkina T, Johnson EA, Hayes PM (2011) Construction and application for QTL analysis of a restriction site associated DNA (RAD) linkage map in barley. BMC Genom 12:4. doi:10.1186/1471-2164-12-4
Das M, Banerjee S, Topdar N, Kundu A, Sarkar D, Sinha MK, Balyan HS, Gupta PK (2011) Development of large-scale AFLP markers in jute. J Plant Biochem Biotechnol 20:270–275. doi:10.1007/s13562-011-0058-1
Das M, Banerjee S, Dhariwal R, Mir RR, Vyas S, Topdar N, Kundu A, Khurana JP, Tyagi AK, Sarkar D, Sinha MK, Balyan HS, Gupta PK (2012a) Development of SSR markers and construction of a linkage map in jute. J Genet 91:21–31. doi:10.1007/s12041-012-0151-9
Das M, Banerjee S, Topdar N, Kundu A, Mir RR, Sarkar D, Sinha MK, Balyan HS, Gupta PK (2012b) QTL identification for molecular breeding of fibre yield and fibre quality traits in jute. Euphytica 187:175–189. doi:10.1007/s10681-011-0603-y
Edwards JH (1991) The Oxford Grid. Ann Hum Genet 55:17–31. doi:10.1111/j.1469-1809.1991.tb00394.x
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379. doi:10.1371/journal.pone.0019379
Fenart S, Ndong Y-P, Duarte J, Riviere N, Wilmer J, van Wuytswinkel O, Lucau A, Cariou E, Neutelings G, Gutierrez L, Chabbert B, Guillot X, Tavernier R, Hawkins S, Thomasset B (2010) Development and validation of a flax (Linum usitatissimum L.) gene expression oligo microarray. BMC Genom 11:592. doi:10.1186/1471-2164-11-592
Fishman L, Willis JH (2005) A novel meiotic drive locus almost completely distorts segregation in Mimulus (Monkeyflower) hybrids. Genetics 169:347–353. doi:10.1534/genetics.104.032789
Gallais A (2009) Hétérosis et variétés hybrides en amelioration des plantes. Quæ, Versailles
Gardner KM, Latta RG (2007) Shared quantitative trait loci underlying the genetic correlation between continuous traits. Mol Ecol 16:4195–4209. doi:10.1111/j.1365-294X.2007.03499.x
Goffinet B, Gerber S (2000) Quantitative trait loci: a meta-analysis. Genetics 155:463–473
Graham GI, Wolff DW, Stuber CW (1997) Characterization of a yield quantitative trait locus on chromosome five of maize by fine mapping. Crop Sci 37:1601–1610. doi:10.2135/cropsci1997.0011183X003700050033x
Guo L, Wang K, Chen J, Huang D, Fan Y, Zhuang J (2013) Dissection of two quantitative trait loci for grain weight linked in repulsion on the long arm of chromosome 1 of rice (Oryza sativa L.). Crop J 1:70–76. doi:10.1016/j.cj.2013.07.008
Hall MC, Willis JH (2005) Transmission ratio distortion in intraspecific hybrids of Mimulus guttatus: implications for genomic divergence. Genetics 170:375–386. doi:10.1534/genetics.104.038653
Hegarty M, Yadav R, Lee M, Armstead I, Sanderson R, Scollan N, Powell W, Skøt L (2013) Genotyping by RAD sequencing enables mapping of fatty acid composition traits in perennial ryegrass (Lolium perenne (L.)). Plant Biotechnol J 11:572–581. doi:10.1111/pbi.12045
Jankauskienė Z, Gruzdevienė E (2013) Physical parameters of dew retted and water retted hemp (Cannabis sativa L.) fibres. Zemdirbyste-Agriculture 100:71–80. doi:10.13080/z-a.2013.100.010
Jaramillo-Correa J, Verdu M, Gonzalez-Martinez S (2010) The contribution of recombination to heterozygosity differs among plant evolutionary lineages and life-forms. BMC Evol Biol 10:22. doi:10.1186/1471-2148-10-22
Kakioka R, Kokita T, Kumada H, Watanabe K, Okuda N (2013) A RAD-based linkage map and comparative genomics in the gudgeons (genus Gnathopogon, Cyprinidae). BMC Genom 14:32. doi:10.1186/1471-2164-14-32
Knapp SJ, Bridges WC Jr (1987) Confidence interval estimators for heritability for several mating and experiment designs. Theor Appl Genet 73:759–763. doi:10.1007/bf00260787
Knapp SJ, Stroup WW, Ross WM (1985) Exact confidence intervals for heritability on a progeny mean basis. Crop Sci 25:192–194. doi:10.2135/cropsci1985.0011183X002500010046x
Kundu A, Sarkar D, Bhattacharjee A, Topdar N, Sinha MK, Mahapatra BS (2011) A simple ethanol wash of the tissue homogenates recovers high-quality genomic DNA from Corchorus species characterized by highly acidic and proteinaceous mucilages. Electron J Biotechnol 14:10–11. doi:10.2225/vol14-issue1-fulltext-4
Kundu A, Sarkar D, Mandal NA, Sinha MK, Mahapatra BS (2012) A secondary phloic (bast) fibre-shy (bfs) mutant of dark jute (Corchorus olitorius L.) develops lignified fibre cells but is defective in cambial activity. Plant Growth Regul 67:45–55. doi:10.1007/s10725-012-9660-z
Kundu A, Topdar N, Sarkar D, Sinha MK, Ghosh A, Banerjee S, Das M, Balyan HS, Mahapatra BS, Gupta PK (2013) Origins of white (Corchorus capsularis L.) and dark (C. olitorius L.) jute: a reevaluation based on nuclear and chloroplast microsatellites. J Plant Biochem Biotechnol 22:372–381. doi:10.1007/s13562-012-0165-7
Larièpe A, Mangin B, Jasson S, Combes V, Dumas F, Jamin P, Lariagon C, Jolivot D, Madur D, Fiévet J, Gallais A, Dubreuil P, Charcosset A, Moreau L (2012) The genetic basis of heterosis: multiparental quantitative trait loci mapping reveals contrasted levels of apparent overdominance among traits of agronomical interest in maize (Zea mays L.). Genetics 190:795–811. doi:10.1534/genetics.111.133447
Matsumura H, Miyagi N, Taniai N, Fukushima M, Tarora K, Shudo A, Urasaki N (2014) Mapping of the gynoecy in bitter gourd (Momordica charantia) using RAD-seq analysis. PLoS One 9:e87138. doi:10.1371/journal.pone.0087138
Miller JR, Koren S, Sutton G (2010) Assembly algorithms for next-generation sequencing data. Genomics 95:315–327. doi:10.1016/j.ygeno.2010.03.001
Mir RR, Rustgi S, Sharma S, Singh R, Goyal A, Kumar J, Gaur A, Tyagi AK, Khan H, Sinha MK, Balyan HS, Gupta PK (2008) A preliminary genetic analysis of fibre traits and the use of new genomic SSRs for genetic diversity in jute. Euphytica 161:413–427. doi:10.1007/s10681-007-9597-x
Mir RR, Banerjee S, Das M, Gupta V, Tyagi AK, Sinha MK, Balyan HS, Gupta PK (2009) Development and characterization of large-scale simple sequence repeats in jute. Crop Sci 49:1687–1694. doi:10.2135/cropsci2008.10.0599
Pfender WF, Saha MC, Johnson EA, Slabaugh MB (2011) Mapping with RAD (restriction-site associated DNA) markers to rapidly identify QTL for stem rust resistance in Lolium perenne. Theor Appl Genet 122:1467–1480. doi:10.1007/s00122-011-1546-3
Ren Y, Zhao H, Kou Q, Jiang J, Guo S, Zhang H, Hou W, Zou X, Sun H, Gong G, Levi A, Xu Y (2012) A high resolution genetic map anchoring scaffolds of the sequenced watermelon genome. PLoS One 7:e29453. doi:10.1371/journal.pone.0029453
Ross AJ, Hallauer AR, Lee M (2006) Genetic analysis of traits correlated with maize ear length. Maydica 51:301–313
Rowe HC, Renaut S, Guggisberg A (2011) RAD in the realm of next-generation sequencing technologies. Mol Ecol 20:3499–3502. doi:10.1111/j.1365-294X.2011.05197.x
Rowell RM, Stout HP (2007) Jute and kenaf. In: Lewin M (ed) Handbook of fibre chemistry, 3rd edn. CRC Press, Boca Raton, pp 405–452
Roy A, Bandyopadhyay A, Mahapatra AK, Ghosh SK, Singh NK, Bansal KC, Koundal KR, Mohapatra T (2006) Evaluation of genetic diversity in jute (Corchorus species) using STMS, ISSR and RAPD markers. Plant Breed 125:292–297. doi:10.1111/j.1439-0523.2006.01208.x
Saha P, Sarkar D, Kundu A, Majumder S, Datta SK, Datta K (2014) Karyotype analysis and chromosomal evolution in Asian species of Corchorus (Malvaceae s. l.). Genet Resour Crop Evol 61:1173–1188. doi:10.1007/s10722-014-0099-0
Sarkar D, Kundu A, Saha A, Mondal NA, Sinha MK, Mahapatra BS (2011) First nuclear DNA amounts in diploid (2n = 2x = 14) Corchorus spp. by flow cytometry: genome sizes in the cultivated jute species (C. capsularis L. and C. olitorius L.) are ~300 % smaller than the reported estimate of 1100–1350 Mb. Caryologia 64:147–153. doi:10.1080/00087114.2002.10589776
Scaglione D, Acquadro A, Portis E, Tirone M, Knapp S, Lanteri S (2012) RAD tag sequencing as a source of SNP markers in Cynara cardunculus L. BMC Genom 13:3. doi:10.1186/1471-2164-13-3
Segonzac C, Boyer J-C, Ipotesi E, Szponarski W, Tillard P, Touraine B, Sommerer N, Rossignol M, Gibrat R (2007) Nitrate efflux at the root plasma membrane: identification of an Arabidopsis excretion transporter. Plant Cell 19:3760–3777. doi:10.1105/tpc.106.048173
Singh A, Rana MK, Singh S, Kumar S, Kumar R, Singh R (2014) CAAT box-derived polymorphism (CBDP): a novel promoter-targeted molecular marker for plants. J Plant Biochem Biotechnol 23:175–183. doi:10.1007/s13562-013-0199-5
Spokevicius AV, Southerton SG, MacMillan CP, Qiu D, Gan S, Tibbits JFG, Moran GF, Bossinger G (2007) β-tubulin affects cellulose microfibril orientation in plant secondary fibre cell walls. Plant J 51:717–726. doi:10.1111/j.1365-313X.2007.03176.x
Swaminathan MS, Iyer RD (1961) Skewed recombination in a rare interspecific jute hybrid. Nature 192:893–894. doi:10.1038/192893b0
Swaminathan MS, Iyer RD, Subbha K (1961) Morphology, cytology and breeding behaviour of hybrids between Corchorus olitorius and C. capsularis. Curr Sci 30:67–68
Topdar N, Kundu A, Sinha MK, Sarkar D, Das M, Banerjee S, Kar CS, Satya P, Balyan HS, Mahapatra BS, Gupta PK (2013) A complete genetic linkage map and QTL analyses for bast fibre quality traits, yield and yield components in jute (Corchorus olitorius L.). Cytol Genet 47:129–137. doi:10.3103/s0095452713030092
Utz HF (2011) PLABSTAT, a computer program for statistical analysis of plant breeding experiments. University of Hohenheim, Stuttgart
van den Broeck HC, Maliepaard C, Ebskamp MJM, Toonen MAJ, Koops AJ (2008) Differential expression of genes involved in C1 metabolism and lignin biosynthesis in wooden core and bast tissues of fibre hemp (Cannabis sativa L.). Plant Sci 174:205–220. doi:10.1016/j.plantsci.2007.11.008
Van Ooijen JW (2006) JoinMap® 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma B. V., Wageningen
Van Ooijen JW (2009) MapQTL® 6, software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma B. V., Wageningen
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. doi:10.1093/jhered/93.1.77
Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L et al (2012a) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44:1098–1103. doi:10.1038/ng.2371
Wang Y-Y, Hsu P-K, Tsay Y-F (2012b) Uptake, allocation and signaling of nitrate. Trends Plant Sci 17:458–467. doi:10.1016/j.tplants.2012.04.006
Ward JA, Bhangoo J, Fernández-Fernández F, Mloore P, Swanson JD, Viola R, Velasco R, Bassil N, Weber CA, Sargent DJ (2013) Saturated linkage map construction in Rubus idaeus using genotyping by sequencing and genome-independent imputation. BMC Genom 14:2. doi:10.1186/1471-2164-14-2
Whitlock BA, Bayer C, Baum DA (2001) Phylogenetic relationships and floral evolution of the Byttnerioideae (“Sterculiaceae” or Malvaceae s.l.) based on sequences of the chloroplast gene, ndhF. System Bot 26:420–437. doi:10.1043/0363-6445-26.2.420
Yang H, Tao Y, Zheng Z, Li C, Sweetingham M, Howieson J (2012) Application of next-generation sequencing for rapid marker development in molecular plant breeding: a case study on anthracnose disease resistance in Lupinus angustifolius L. BMC Genom 13:318. doi:10.1186/1471-2164-13-318
Acknowledgments
We acknowledge the Xcelris Labs Ltd., Ahmedabad, for assistance in Illumina HiSeq 2000 sequencing, raw data processing and the related bioinformatics analyses. We thank Director, Sugarcane Breeding Institute (SBI), Coimbatore, for allowing us to advance the mapping population in the off-season nursery, Dr. N. Subramonian (SBI, Coimbatore) for extending laboratory facilities for DNA isolation and Surajeet Dey (CRIJAF, Barrackpore) for field assistance in the development and maintenance of the mapping populations. The research was funded by National Fund for Basic, Strategic and Frontier Application Research in Agriculture (NFBSFARA), Indian Council of Agricultural Research (ICAR), New Delhi (F. No. NFBSFARA/GB-2018/2011-12), and ICAR Network Project of Transgenics in Crops (ICAR-NPTC), ICAR, New Delhi (F. No. ICAR/NRCPB/NPTC-3052). Comments and suggestions on the manuscript from two anonymous reviewers are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kundu, A., Chakraborty, A., Mandal, N.A. et al. A restriction-site-associated DNA (RAD) linkage map, comparative genomics and identification of QTL for histological fibre content coincident with those for retted bast fibre yield and its major components in jute (Corchorus olitorius L., Malvaceae s. l.). Mol Breeding 35, 19 (2015). https://doi.org/10.1007/s11032-015-0249-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0249-x