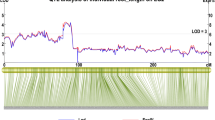
Abstract
Binary traits are often encountered in plant (and animal) breeding. These include resistance/susceptibility to diseases or the presence/absence of a given characteristic. Root vigor in sugar beet is related to nutrient uptake from the soil and sugar yield and can be classified as either high or low, thus providing an example of binary trait of agronomic importance. Genomic data may be used for early prediction of seedlings root vigor in sugar beet breeding programmes. In this context, it may be of theoretical and practical interest to determine the minimum set of data needed for accurate predictions. A panel of 175 SNP markers was used to genotype 123 sugar beet individual plants. SNPs were ranked based on their predictive ability in a model selection algorithm. Starting from the bottom (least relevant SNP), one SNP at a time was removed and the predictive ability of the remaining SNPs assessed. The accuracy of prediction was in general very high, close to 100 %. Only starting from \(\le\)30 SNPs in the model, the prediction accuracy became less stable and began to decrease. Based on results, a set of 30–50 SNPs can be recommended for accurate prediction of root vigor in sugar beet populations. The described procedure is in principle applicable to any binary trait in any plant (or animal) species of agricultural interest.





Similar content being viewed by others
References
Abou-Elwafa S, Büttner B, Kopisch-Obuch F, Jung C, Müller A (2012) Genetic identification of a novel bolting locus in Beta vulgaris which promotes annuality independently of the bolting gene B. Mol Breed 29:989–998
Biancardi E, McGrath JM, Panella LW, Lewellen RT, Stevanato P (2010) Sugar beet. In: Bradshaw JE (ed) Root and tuber crops. Springer, Berlin, pp 173–219
Biscarini F, Stevanato P, Broccanello C, Stella A, Saccomani M (2014) Genome-enabled predictions for binomial traits in sugar beet populations. BMC Genet 18(5):1–9
Chaitin G (2006) The limits of reason. Sci Am 294(3):74–81
Dohm JC, Minoche AE, Holtgräwe D, Capella-Gutiérrez S, Zakrzewski F, Tafer H, Rupp O, Sörensen TR, Stracke R, Reinhardt R et al (2013) The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505:546–549
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PloS One 6(5):e19,379
FAO Statistics Division (2012) Worldwide centrifuged raw sugar production in year 2012. Retrieved from the FAOSTAT database. http://faostat.fao.org/site/636/DesktopDefault.aspx?PageID=636
Fawcett T (2004) Roc graphs: notes and practical considerations for researchers. Mach Learn 31:1–38
Foulongne M, Pascal T, Pfeiffer F, Kervella J (2003) QTLs for powdery mildew resistance in peach\(\times\) Prunus davidiana crosses: consistency across generations and environments. Mol Breed 12(1):33–50
Georges M (2014) Towards sequence-based genomic selection of cattle. Nat Genet 46(8):807–809
Gianola D, de Los Campos G, Hill WG, Manfredi E, Fernando R (2009) Additive genetic variability and the bayesian alphabet. Genetics 183(1):347–363
Goddard M, Hayes B (2007) Genomic selection. J Anim Breed Genet 124(6):323–330
Gu XY, Foley ME, Horvath DP, Anderson JV, Feng J, Zhang L, Mowry CR, Ye H, Suttle JC, Kadowaki Ki (2011) Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice. Genetics 189(4):1515–1524
Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH (2009) The weka data mining software: an update. ACM SIGKDD Explor Newsl 11(1):10–18
Hastie T, Tibshirani R, Friedman J (2009) Model assessment and selection. In: Casella G, Fienberg S, Olkin I (eds) The elements of statistical learning. Springer, New York, pp 219–260
Heffner EL, Lorenz AJ, Jannink JL, Sorrells ME (2010) Plant breeding with genomic selection: gain per unit time and cost. Crop Sci 50(5):1681–1690
Hofheinz N, Borchardt D, Weissleder K, Frisch M (2012) Genome-based prediction of test cross performance in two subsequent breeding cycles. Theor Appl Genet 125(8):1639–1645
Jonas E, de Koning DJ (2013) Does genomic selection have a future in plant breeding? Trend Biotechnol 31(9):497–504
Karegowda AG, Manjunath A, Jayaram M (2010) Comparative study of attribute selection using gain ratio and correlation based feature selection. Int J Inf Technol Knowl Manag 2(2):271–277
Liu Z, Shen Y, Ott J (2011) Multilocus association mapping using generalized ridge logistic regression. BMC Bioinfo 12(1):384
Lorenz A, Smith K, Jannink JL (2012) Potential and optimization of genomic selection for fusarium head blight resistance in six-row barley. Crop Sci 52(4):1609–1621
MATLAB (2010) version 7.10.0 (R2010a). The MathWorks Inc., Natick, MA
Meuwissen T, Hayes B, Goddard (2001) Goddard: prediction of total genetic value using genome-wide dense marker maps. Genetics 157(4):1819–1829
Moose SP, Mumm RH (2008) Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol 147(3):969–977
Nicolazzi EL, Picciolini M, Strozzi F, Schnabel RD, Lawley C, Pirani A, Brew F, Stella A (2014) SNPchiMp: a database to disentangle the SNPchip jungle in bovine livestock. BMC Genomics 15(1):123
Quinlan JR (1986) Induction of decision trees. Mach Learn 1(1):81–106
R Development Core Team (2008) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org. ISBN 3-900051-07-0
Russu A, Malovini A, Puca AA, Bellazzi R (2012) Stochastic model search with binary outcomes for genome-wide association studies. J Am Med Info Assoc 19(e1):e13–e20
Stevanato P, Broccanello C, Biscarini F, Del Corvo M, Sablok G, Panella L, Stella A, Concheri G (2013) High-throughput RAD-SNP genotyping for characterization of sugar beet genotypes. Plant Mol Biol Rep 32(3):691–696
Stevanato P, Saccomani M, Bertaggia M, Bottacin A, Cagnin M, De Biaggi M, Biancardi E (2004) Nutrient uptake traits related to sugarbeet yield. J Sugar Beet Res 41:89–100
Stevanato P, Trebbi D, Saccomani M (2010) Root traits and yield in sugar beet: identification of AFLP markers associated with root elongation rate. Euphytica 173(3):289–298
Turner EH, Ng SB, Nickerson DA, Shendure J (2009) Methods for genomic partitioning. Annu Rev Genomics Hum Genet 10:263–284
Wang C, Ding X, Wang J, Liu J, Fu W, Zhang Z, Yin Z, Zhang Q (2013) Bayesian methods for estimating GEBVs of threshold traits. Heredity 110(3):213–219
Würschum T, Reif JC, Kraft T, Janssen G, Zhao Y (2013) Genomic selection in sugar beet breeding populations. BMC Genet 14(1):85
Yang Y, Pedersen JO (1997) A comparative study on feature selection in text categorization. ICML 97:412–420
Acknowledgments
This research was financially supported by the Marie Curie European Reintegration Grant “NEUTRADAPT.”
Author information
Authors and Affiliations
Corresponding author
Additional information
Filippo Biscarini and Nelson Nazzicari have contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Biscarini, F., Marini, S., Stevanato, P. et al. Developing a parsimonius predictor for binary traits in sugar beet (Beta vulgaris). Mol Breeding 35, 10 (2015). https://doi.org/10.1007/s11032-015-0197-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0197-5