Abstract
Exosomes are nanometer-sized vesicles involved in intercellular communication, and they are released by various cell types. To learn about exosomes produced by Schwann cells (SCs) and to explore their potential function in repairing the central nervous system (CNS), we isolated exosomes from supernatants of SCs by ultracentrifugation, characterized them by electron microscopy and immunoblotting and determined their protein profile using proteomic analysis. The results demonstrated that Schwann cell-derived exosomes (SCDEs) were, on average, 106.5 nm in diameter, round, and had cup-like concavity and expressed exosome markers CD9 and Alix but not tumor susceptibility gene (TSG) 101. We identified a total of 433 proteins, among which 398 proteins overlapped with the ExoCarta database. According to their specific functions, we identified 12 proteins that are closely related to CNS repair and classified them by different potential mechanisms, such as axon regeneration and inflammation inhibition. Gene Oncology analysis indicated that SCDEs are mainly involved in signal transduction and cell communication. Biological pathway analysis showed that pathways are mostly involved in exosome biogenesis, formation, uptake and axon regeneration. Among the pathways, the neurotrophin, PI3K-Akt and cAMP signaling pathways played important roles in CNS repair. Our study isolated SCDEs, unveiled their contents, presented potential neurorestorative proteins and pathways and provided a rich proteomics data resource that will be valuable for future studies of the functions of individual proteins in neurodegenerative diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Central nervous system (CNS) injury, such as spinal cord injury and brain injury, causes irreversible loss of motor and sensory function [1, 2]. Due to the development of modern society, the occurrence rate of CNS damage increases year by year. The predicament of CNS regeneration is always attributed to the poor regenerative plasticity of mature neurons as well as the detrimental microenvironment caused by the lesion [3]. The peripheral nervous system (PNS) exhibits adequate regeneration after injury, which is different from the CNS. During this process, glial cells in situ play a more important role than functional neurons. In the CNS, multiple growth inhibitory factors derived from oligodendrocytes, such as Nogo, myelin-associated glycoprotein (MAG), and oligodendrocyte myelin glycoprotein (OMgp), induce the collapse of the growth cone [4]. Conversely, in the PNS, Schwann cells promote nerve regeneration through secreting growth factors, clearing myelin and axonal debris, activating macrophages and forming new medullary sheath [5].
Taking the advantages of Schwann cells into consideration, many studies have attempted to transplant these cells into injured CNS. This strategy obtained many encouraging results [6]. The advantages of Schwann cell transplantation include the practicability of autologous transplantation [7, 8] and synergistic effects combined with other strategies [9, 10]. However, some evidence still causes researchers to believe that Schwann cells transplanted into the CNS cannot perform as ideally as they can in the PNS. A major challenge for Schwann cell CNS transplantation is the low graft cell survival due to p75NTR-induced apoptosis [11, 12] and limited migration ability across the astrocyte boundary [13, 14]. Therefore, whether a previous favorable improvement is the mutual factor between active and adverse effects after SC transplantation in the CNS remains to be determined. We hypothesized that there should be a new strategy to optimize the ability of SCs to minimize undesirable characteristics.
Exosomes are small vesicles with a diameter of approximately 50–100 nm that are produced in the endosomal compartment and filled with functional proteins, microRNAs (miRNAs) and mRNAs [15]. They are involved in intercellular communication and are secreted continuously under many physiological and pathological conditions [16]. Their subparticle nature in the host cells makes them serve as microenvironment modulators through paracrine mechanisms to ease stimuli [17]. A recent study demonstrated that exosomes derived from Schwann cells support axonal maintenance and regeneration after PNS damage [18]. Considering the positive performance of SCs in CNS repair, we wondered whether the use of their exosomes is the optimal strategy to replace cell transplantation. Because of the complexity CNS composition, understanding exosome content is necessary to explore potential target cells. Until now, only individual proteins have been identified by various studies [19], which leads to the requirement of comprehensive information about the proteins in Schwann cell-derived exosomes (SCDEs).
In the current study, we purified exosomes from SC-conditioned medium and explored their morphological features and biomarkers. Furthermore, 433 proteins were identified from SCDEs using LC-MS/MS. Systematic proteomics characteristics were obtained through bioinformatics analysis. This study provides new evidence that SCDEs may act as a novel therapeutic strategy for CNS injury.
Methods
Isolation of exosome vesicles by ultracentrifugation
Primary SCs derived from the sciatic nerve of adult Wistar rats (female, 230 g ± 10 g, provided by the Academy of Military Medical Sciences, Tianjin, China) were cultured, and exosome isolation was performed as described previously with minor modifications in Fig. 1 [9, 18]. All animal procedures were approved by the Ethics Committee of Tianjin Medical University and were in accordance with the Guidance Suggestions for the Care and Use of Laboratory Animals. First, the SCs were cultured in exosome-depleted growth medium, and the cell conditioned medium was collected. Then, dead cells and large growth debris were removed through centrifugation (1000 × g, 20 min). Second, to remove additional debris, the supernatant was centrifuged at 10,000 × g for 30 min. Then, the supernatant was filtered using 0.22-µm filters (Millipore, CA). Then, the filtered supernatant was centrifuged at 100,000 × g for 70 min to collect the exosomes. Finally, the exosomes were resuspended in cold PBS for further experiments.
Characterization of SCDEs by transmission electron microscopy (TEM) and the Malvern Zetasizer Nano ZS90
Transmission electron microscopy (TEM) was used for morphological observation. The exosome samples were prepared as described above. For TEM, briefly, the exosomes were fixed with 2.5% glutaraldehyde overnight at 4 °C. The solution was centrifuged at 100,000 × g to remove the glutaraldehyde, and the exosomes were washed three times with PBS. Then, the exosomes were stained with 3% phosphotungstic acid aqueous solution and fixed on copper mesh formvar grids. A transmission electron microscope (JEM-1010) was used to detect the exosomes. A Malvern Zetasizer Nano ZS90 (Malvern, UK) was used to detect the concentration of exosomes in different samples. Samples with appropriate concentrations were used to examine the size distribution of exosomes.
Western blot
Exosome samples were lysed with RIPA buffer, and the collected protein samples were added to 5 × SDS loading buffer and denatured by boiling for 5 min. Then, 10% acrylamide gels were used for electrophoresis, and the proteins were transferred to PVDF membranes for 2 h. After washing three times with TBST, the membranes were blocked with blocking buffer (5% nonfat milk in PBS) for 1 h at room temperature (RT). Then, the membranes were incubated with primary antibodies overnight at 4 °C. The antibodies were diluted as follows: CD9 (rabbit monoclonal 1:2000; Abcam), Alix (mouse monoclonal 1:1000; Cell Signaling Technology) and TSG101 (rabbit monoclonal 1:500; Abcam). The next day, the membranes were washed three times with TBST and incubated with secondary antibody (1:2000) for 1 h at RT. Blots were detected using enhanced chemiluminescence.
Protein digestion
Briefly, the obtained exosomes were lysed by 4% SDS, 100 mM DTT, and 50 mM Tris, and HCl (pH 7.5). According to the FASP protocol, trypsin was used to digest total proteins (20 µg). The proteins were denatured for 5 min at 95 °C and mixed with 200 mL UA buffer solution (8 M urea in 0.1 M Tris HCl pH 8.5) and centrifuged (12,000 rpm) three times for 15 min. Next, the samples and 100 mL of iodoacetamide (IAA) were mixed at 600 rpm in a hot mixer. The filter unit was incubated for 20 min and centrifuged for 10 min at 12,000 rpm. One hundred milliliters of UA was added to the filter unit and centrifuged at 12,000 rpm three times for 15 min. Then, 100 mL of 50 mM NH4HCO3 was added to the filter unit and centrifuged at 12,000 rpm three times for 15 min. Then, the samples were mixed with trypsin (with 50 mM NH4HCO3) and placed in the thermal mixer at 600 rpm. Then, these units were incubated in a wet chamber at 37 °C overnight. The filtration unit was transferred into the new collecting pipe and centrifuged for 15 min at 12,000 rpm. Then, 50 mM NH4HCO3 was added to the filter unit and centrifuged for 15 min at 12,000 rpm. Then, the samples were dried and stored at − 20 °C until LC-MS/MS.
LC-MS/MS
Referring to a previous study [20], LC-MS/MS was performed with some modifications. In short, the final concentration of the samples was 0.1% (V/V) after dilution with formic acid. The samples were then loaded onto a 75 mm x 150 mm fused silica column that was packed in-house with 3-mm ReproSil-Pur C18 beads (120 Å; Dr. Maisch GmbH, Ammerbuch, Germany). LC-MS/MS was performed using an Easy Nano-UPLC 1000 (Thermo Electron, Waltham, MA). A gradient (5–80% acetonitrile and 0.1% formic acid) was used to elute peptides on the Q-Exactive mass spectrometer (Thermo Electron, Waltham, MA) in the flow rate of 300 l/min over 240 min. The MS/MS spectrum was obtained in the data-dependent mode, and the scan resolution was 70,000 during acquisition (m/z 200).
Data processing
Raw data files were processed with Proteome Discoverer (v1.3; Thermo Scientific).
Then, Mascot 2.3.02 (Matrix Science) Boston, MA, software was used to search against the Bovine RefSeq database. Two missed cleavages are allowed for tryptic enzymes. Carbamidomethylation (C) was set to fixed modification, and oxidation (M) and acetylation (N-term) were set as variable modifications. The peptide difference was 20 ppm, the fragment ion mass error was 0.1 Da, and the peptide and protein identification error rates were < 1%.
Gene Ontology annotation and pathway analysis
Functional-enrichment analysis for Gene Ontology (GO) terms was conducted through the Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.8) (https://david.ncifcrf.gov/) with the entire murine genome as the background. The enriched GO analysis of annotated proteins was performed for cellular components, molecular functions and biological processes. Pathways enriched with the proteins were generated by KEGG pathway analysis and Cytoscape software 3.5.1. The Venn diagrams web tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to compare the proteins identified with the ExoCarta database. GraphPad Prism 6.0 was used for plotting.
Results
Characterizations of Schwann cell-derived exosomes
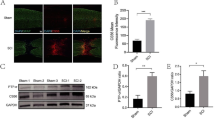
To explore the neuroprotective effect of SCDEs, we isolated exosomes from cultured Schwann cells. Schwann cells were positive for S100 (Fig. 2a). After observation using the Malvern Zetasizer Nano ZS90, the mean diameter of the SCDEs was 106.5 nm (Fig. 2b). We performed Western blotting to detect CD9, Alix and TSG101 expression to characterize these exosomes. The results showed that CD9 and Alix, which are exosome markers, could be detected, but TSG101 was not (Fig. 2c), which is consistent with exosome characteristics [18]. In addition, the morphology of SC-released exosomes was further confirmed through TEM, which showed SCDEs with a size range of 40–100 nm (Fig. 2d).
Characterizations of Schwann cell-derived exosomes. Characterization of the isolated SCDEs. a Schwann cells were positive for S100. (Bar = 50 µm). b The size distribution of SCDEs was detected by the Malvern Zetasizer Nano ZS90. c The expression of CD9, Alix, and TSG101 was detected by Western blot in the Schwann cell lysate (SCL) group and Schwann cell-derived exosome (SCDE) group. d TEM image of SCDEs
Proteomic analysis of Schwann cell-derived exosomes
We repeated the experiments three times (Supplementary Figure). To reveal the mechanisms of the neuroprotective effect of SCDEs, we performed proteomic analysis on SCDEs. Finally, 433 proteins in exosomes that were concordant in the three biological duplicates were identified and compared with the exosome database, ExoCarta; 398 proteins overlapped (Fig. 3) and were ranked based on intensity values (Supplementary Table). Thus, 91.92% of the identified proteins overlapped with ExoCarta, which indicated that the procedures for isolation and purification are repeatable, and the results of proteomic analysis are reliable. As shown in Table 1, most of the identified proteins are related to CNS repair.
SCDEs might enhance axon regeneration
Several studies have reported that SCDEs play a critical role in axonal regeneration in the PNS [18, 44, 45]. However, whether SCDEs enhance axon regeneration in the CNS is still unclear. According to the proteomics results, twelve proteins were closely related to axon regeneration, such as carboxypeptidase E (CPE), fatty acid-binding protein (FABP5), fibronectin, flotillin-2, major vault protein (MVP), monocarboxylate transporter 1 (MCT1), neuropilin-2 (NRP2), septin-7 (SEPT7), protein disulfide-isomerase A3 (PDIA3) and syntenin-1. As SCDEs contain proteins involved in axon regeneration, this result reveals that the function of SCs in promoting axonal regeneration might be through exosomes.
SCDEs might inhibit inflammation
Exosomes from several kinds of cells participate in the inhibition of the inflammatory response, which is considered a novel therapeutic approach for some diseases [46,47,48]. Likewise, two proteins that we found in SCDEs, αB-crystallin and galectin-1, might produce benefits similar to anti-inflammatory effects in CNS injury. Thus, SCDEs might have an anti-inflammatory role in CNS damage.
Functional categories
Next, GO annotation was performed to determine the functional roles of the SCDE proteins (SCDEP) via DAVID version 6.8 and to obtain the enriched terms for molecular function, biological process and cellular component. The GO classification system revealed that the proteins could be classified into groups according to their functional properties (Fig. 4a–c). The biological processes analysis revealed enrichment of SCDEP related to “cell adhesion”, “negative regulation of apoptotic” and “signal transduction”. For the cellular component, these proteins were enriched in “exosomes”, the “cytoplasm” and the “membrane”. The molecular functions of the exosome proteins are mainly enriched in “protein binding”, “poly (A) RNA binding” and “GTP binding”.
KEGG pathway analysis
The enriched signaling pathways with the exosome proteins were determined using KEGG pathway analysis. The top 20 pathways based on enrichment were given. As shown in Table 2, the top 20 pathways enriched with a p value < 0.05 were focal adhesion, endocytosis, regulation of the actin cytoskeleton and the PI3K-Akt signaling pathway. Among these, the neurotrophin signaling pathway, PI3K-Akt signaling pathway and cAMP signaling pathway are related to the CNS microenvironment.
Discussion
Schwann cells are the main functional glial cells in the PNS and play a very important role in axon regeneration after PNS injury. Recent studies believe that Schwann cells and their encapsulated axons should be deemed a “functional syncytium” [44] because of the inadequate capability of overlong axons to transport enough proteins in time and the evidence of substance exchange between axons and Schwann cells. Exosomes are small vesicles carrying functional molecules from host cells and play an important role in cell–cell communication [49]. Exosomes derived from Schwann cells modulate the damaged PNS microenvironment and enhance axonal regeneration by inhibiting the activity of the GTPase RhoA [18]. This finding gives us a new promising strategy for other neurodegenerative disorders, such as amyotrophic lateral sclerosis (ALS), Alzheimer’s disease (AD) and brain and spinal cord injury. Therefore, there is an urgent need to investigate exosome contents, which can help us gain a deeper understanding of potential therapeutic mechanisms. In this study, we isolated SCDEs and verified the size, morphology and surface markers associated with exosome characteristics. Then, proteomics analysis was performed, and 433 proteins were identified through LC-MS/MS. Ultimately, bioinformatics analysis was performed to reveal their potential mechanism.
The Schwann cells in this study were isolated from the sciatic nerves of rats according to our previous methods, and by doing so, the purity of Schwann cells was more than 95% [50]. Until now, there has been no advanced method to purify cultured Schwann cells to 100%, and fibroblast contamination is difficult to avoid [51]. Despite the existence of the infinite cell line RSC96, which acts as an alternative for primary Schwann cells in some aspects [52], comparative proteomic analysis confirmed that this cell line exhibited different secreted protein expression profiles [53]. In this study, we purified Schwann cells to approximately 100% purity, which can give us reliable results.
The GO analysis of proteomics data revealed that the proteins in SCDEs were enriched in cellular processes, cellular processes involving biological regulation, which indicated that the contents of exosomes share close ties with microenvironment regulation. This regulatory characteristic is consistent with the supporting cell status for Schwann cells [54]. In the cellular component of GO analysis, the enrichment of cytoplasm and cytosol reflects the origin of exosomes. In addition, membrane-bound vesicles and plasma membranes are consistent with exosome features. For molecular function, protein binding is the most common characteristic, revealing that direct regulation of protein–protein interaction might be the main kind of regulation for SCDEs.
We used the KEGG pathway database to explore the enrichment of proteins and found that several signaling pathways were related to proteins in SCDEs. Some pathways reflect the basic biofunction of cells, such as the proteasome, glycolysis and regulation of the actin cytoskeleton, which proved that exosomes act as a suborgan of the host cells. We noticed that some proteins were enriched in the ribosome pathway, which is consistent with the fact that Schwann cells can transfer ribosomes to nearby axons and confirms that Schwann cells play an important role in supporting local protein synthesis in axons [55]. In the future, we will explore whether regeneration-related mRNAs exist in such exosomes.
Considering the potential therapeutic ability of CNS, we found that several enriched pathways are related to the CNS microenvironment, including the neurotrophin signaling pathway, PI3K-Akt signaling pathway and cAMP signaling pathway. In the neurotrophin signaling pathway, we found that Rho GTPases were detected, especially Rac1 and Cdc42. The Rho family of GTPases belongs to the Ras superfamily and plays an important role in neuronal development, neuronal survival and neurodegeneration [56]. In the CNS, Rac1 and Cdc42 can promote neurite outgrowth and stimulate regeneration [57]. In contrast to neurons, astrocytes are always activated and form glial scars to inhibit axon regeneration [58]. The high expression of Rac1 can inhibit astrocyte outgrowth [59]. Meanwhile, Rac1 and Cdc42 can cause astrocyte apoptosis induced by neurotoxicity [60]. In addition, activation of Rac1 and Cdc42 can change microglia into the M2-like phenotype, which is beneficial in CNS regeneration [61]. Analogously, Rac1/Cdc42 signaling can promote the migration of oligodendrocytes, the key cells in CNS for myelinization [62]. A recent study also proved that the activation of Rac1 can preserve blood-spinal cord barrier (BSCB) integrity and improve functional recovery after spinal cord injury through protection of endothelial cells [63].
Conclusions
The proteins in SCDEs can create a more permissive microenvironment related to each component in the CNS for regeneration. Our proteomics analysis may provide a novel therapeutic strategy for CNS injury.
References
Ning GZ, Wu Q, Li YL, Feng SQ (2012) Epidemiology of traumatic spinal cord injury in Asia: a systematic review. J Spinal Cord Med 35:229–239. https://doi.org/10.1179/2045772312y.0000000021
Rubiano AM, Carney N, Chesnut R, Puyana JC (2015) Global neurotrauma research challenges and opportunities. Nature 527:S193–S197. https://doi.org/10.1038/nature16035
O’Shea TM, Burda JE, Sofroniew MV (2017) Cell biology of spinal cord injury and repair. J Clin Invest 127:3259–3270. https://doi.org/10.1172/jci90608
Yiu G, He Z (2006) Glial inhibition of CNS axon regeneration. Nat Rev Neurosci 7:617–627. https://doi.org/10.1038/nrn1956
Assinck P, Duncan GJ, Hilton BJ, Plemel JR, Tetzlaff W (2017) Cell transplantation therapy for spinal cord injury. Nat Neurosci 20:637–647. https://doi.org/10.1038/nn.4541
Wiliams RR, Bunge MB (2012) Schwann cell transplantation: a repair strategy for spinal cord injury? Prog Brain Res 201:295–312. https://doi.org/10.1016/b978-0-444-59544-7.00014-7
Zhou XH, Ning GZ, Feng SQ, Kong XH, Chen JT, Zheng YF, Ban DX, Liu T, Li H, Wang P (2012) Transplantation of autologous activated Schwann cells in the treatment of spinal cord injury: six cases, more than five years of follow-up. Cell Transpl 21(Suppl 1):S39
Anderson KD, Guest J, Dietrich WD, Bunge MB, Curiel R, Dididze M, Green BA, Khan A, Pearse DD, Saraf-Lavi E (2017) Safety of autologous human schwann cell transplantation in subacute thoracic spinal cord injury. J Neurotrauma 34(21):2950–2963
Feng S, Kong X, Guo S, Chen J, Pei W, Xinlong MA, Sun Z (2005) Treatment of spinal cord injury with Co-grafts of genetically modified schwann cells by NGF gene and fetal spinal cord cell suspension in the rat. Neurotox Res 7:169–177
Kanno H, Pressman Y, Moody A, Berg R, Muir EM, Rogers JH, Ozawa H, Itoi E, Pearse DD, Bunge MB (2014) Combination of engineered Schwann cell grafts to secrete neurotrophin and chondroitinase promotes axonal regeneration and locomotion after spinal cord injury. J Neurosci 34:1838–1855. https://doi.org/10.1523/jneurosci.2661-13.2014
Masoumeh F, Farzaneh S, Abdolkhaleg D, Zahra Hassannejad P, Fatemeh P, Vafa RM (2011) Schwann cell apoptosis and p75(NTR) siRNA. Iran J Allergy Asthma Immunol 10:53–59
Ahmad I, Fernando A, Gurgel R, Clark JJ, Xu L, Hansen MR (2015) Merlin status regulates p75 NTR expression and apoptotic signaling in Schwann cells following nerve injury ☆. Neurobiol Dis 82:114–122
Grimpe B, Pressman Y, Bunge MB, Silver J (2005) The role of proteoglycans in Schwann cell/astrocyte interactions and in regeneration failure at PNS/CNS interfaces. Mol Cell Neurosci 28:18–29
Mousumi G, Tuesta LM, Rocio P, Samik P, Kiara M, Abderrahman EM, Urs R, Damien Daniel P (2012) Extensive cell migration, axon regeneration, and improved function with polysialic acid-modified Schwann cells after spinal cord injury. Glia 60:979–992
Théry C (2011) Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep 3:15
Mathivanan S, Ji H, Simpson RJ (2010) Exosomes: extracellular organelles important in intercellular communication. J Proteom 73:1907–1920
Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ (2010) Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteom Mcp 9:197
María Alejandra LV, Frederic P, Court FA (2013) Schwann cell-derived exosomes enhance axonal regeneration in the peripheral nervous system. Glia 61:1795–1806
Lopez-Verrilli MA, Court FA (2012) Transfer of vesicles from schwann cells to axons: a novel mechanism of communication in the peripheral nervous system. Frontiers Physiol 3:205
Mathias RA, Lim JW, Ji H, Simpson RJ (2009) Isolation of extracellular membranous vesicles for proteomic analysis. Methods Mol Biol 528:227–242. https://doi.org/10.1007/978-1-60327-310-7_16
Allen GW, Liu JW, León M De (2000) Depletion of a fatty acid-binding protein impairs neurite outgrowth in PC12 cells. Brain Res Mol Brain Res 76:315–324
Cheng Y, Cawley NX, Loh YP (2014) Carboxypeptidase E (NF-α1): a new trophic factor in neuro- protection. Neurosci Bull 30:692–696
King VR, Alovskaya A, Wei DYT, Brown RA, Priestley JV (2010) The use of injectable forms of fibrin and fibronectin to support axonal ingrowth after spinal cord injury. Biomaterials 31:4447–4456
Figueroa JD, Serrano-Illan M, Licero J, Cordero K, Miranda JD, De Leon M (2016) Fatty acid binding protein 5 modulates docosahexaenoic acid-induced recovery in rats undergoing spinal cord injury. J Neurotrauma 33:1436–1449. https://doi.org/10.1089/neu.2015.4186
King VR, Phillips JB, Hunt-Grubbe H, Brown R, Priestley JV (2006) Characterization of non-neuronal elements within fibronectin mats implanted into the damaged adult rat spinal cord. Biomaterials 27:485–496
Koch JC, Solis GP, Bodrikov V, Michel U, Haralampieva D, Shypitsyna A, Tönges L, Bähr M, Lingor P, Stuermer CAO (2013) Upregulation of reggie-1/flotillin-2 promotes axon regeneration in the rat optic nerve in vivo and neurite growth in vitro. Neurobiol Dis 51:168–176
Stuermer CA (2011) Microdomain-forming proteins and the role of the reggies/flotillins during axon regeneration in zebrafish
Hong-Chao P, Jin-Fei L, Li-Ping M, Yan-Qin S, Melitta S (2013) Major vault protein promotes locomotor recovery and regeneration after spinal cord injury in adult zebrafish. Eur J Neurosci 37:203–211
Lu Z, Yan S, Tao W, Mei C, Zheng W, Hong Z, Qiang D (2014) Curcumin promotes neurite outgrowth via reggie-1/flotillin-2 in cortical neurons. Neurosci Lett 559:7–12
Morrison BM, Tsingalia A, Vidensky S, Lee Y, Jin L, Farah MH, Lengacher S, Magistretti PJ, Pellerin L, Rothstein JD (2015) Deficiency in monocarboxylate transporter 1 (MCT1) in mice delays regeneration of peripheral nerves following sciatic nerve crush. Exp Neurol 263:325–338. https://doi.org/10.1016/j.expneurol.2014.10.018
Yong Y, Melitta S (2013) Syntenin-a promotes spinal cord regeneration following injury in adult zebrafish. Eur J Neurosci 38:2280–2289
Zhang M, Ma Z, Qin H, Yao Z (2017) Monocarboxylate transporter 1 in the medial prefrontal cortex developmentally expresses in oligodendrocytes and associates with neuronal amounts. Mol Neurobiol 54:1–12
Makio T, Kenta Y, Atsushi T, Ikuo M, Atsu A, Valérie C, Fujio M (2013) Role of neuropilin-2 in the ipsilateral growth of midbrain dopaminergic axons. Eur J Neurosci 37:1573–1583
Murthy SRK, Thouennon E, Li W-S, Cheng Y, Bhupatkar J, Cawley NX, Lane M, Merchenthaler I, Loh YP (2013) Carboxypeptidase E protects hippocampal neurons during stress in male mice by up-regulating prosurvival BCL2 protein expression. Endocrinology 154:3284–3293
Botia B, Seyer D, Ravni A, Bénard M, Falluel-Morel A, Cosette P, Jouenne T, Fournier A, Vaudry H, Gonzalez BJ (2008) Peroxiredoxin 2 is involved in the neuroprotective effects of PACAP in cultured cerebellar granule neurons. J Mol Neurosci 36:61–72
Zhang S, Wu D, Wang J, Wang Y, Wang G, Yang M (2013) Stress protein expression in early phase spinal cord ischemia/reperfusion injury. Neural Regen Res 8:2225–2235
Claudio H, Milene RC, Sébastien WL, Sonia C, Elisabeth VK, Kinsey M, Joaquín C, Claudio S (2005) The disulfide isomerase Grp58 is a protective factor against prion neurotoxicity. J Neurosci 25:2793–2802
Hetz CA, Claudio S (2006) Stressing out the ER: a role of the unfolded protein response in prion-related disorders. Curr Mol Med 6:37–43
Bracchi-Ricard V, Lambertsen KL, Ricard J, Nathanson L, Karmally S, Johnstone J, Ellman DG, Frydel B, Mctigue DM, Bethea JR (2013) Inhibition of astroglial NF-kappaB enhances oligodendrogenesis following spinal cord injury. J Neuroinflamm 10:869
Gan Y, Ji X, Hu X, Luo Y, Zhang L, Li P, Liu X, Yan F, Vosler P, Gao Y, Stetler RA, Chen J (2012) Transgenic overexpression of peroxiredoxin-2 attenuates ischemic neuronal injury via suppression of a redox-sensitive pro-death signaling pathway. Antioxid Redox Signal 17:719–732. https://doi.org/10.1089/ars.2011.4298
Hua H, Yayi X, Shuanke W, Bin Z, Zhengyi S, Lingwei Y (2011) Synergistic effects of galectin-1 and reactive astrocytes on functional recovery after contusive spinal cord injury. Arch Orthop Trauma Surg 131:829–839
Blackmore M, Letourneau PC (2006) L1, beta1 integrin, and cadherins mediate axonal regeneration in the embryonic spinal cord. J Neurobiol 66:1564–1583. https://doi.org/10.1002/neu.20311
Parri M, Chiarugi P (2010) Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal 8:23. https://doi.org/10.1186/1478-811x-8-23
Lopez-Leal R, Alvarez J, Court FA (2016) Origin of axonal proteins: Is the axon-schwann cell unit a functional syncytium? Cytoskeleton (Hoboken) 73:629–639. https://doi.org/10.1002/cm.21319
Lopez-Leal R, Court FA (2016) Schwann cell exosomes mediate neuron–glia communication and enhance axonal regeneration. Cell Mol Neurobiol 36:429–436
Seon-Hee K, Lechman ER, Nicole B, Rajasree M, Annahita K, Joan N, Zhibao M, Watkins SC, Andrea G, Robbins PD (2005) Exosomes derived from IL-10-treated dendritic cells can suppress inflammation and collagen-induced arthritis. J Immunol 174:6440
Seon Hee K, Bianco NR, Shufesky WJ, Morelli AE, Robbins PD (2007) MHC class II + exosomes in plasma suppress inflammation in an antigen-specific and Fas ligand/Fas-dependent manner. J Immunol 179:2235–2241
Teng H, Hu M, Yuan LX, Liu Y, Guo X, Zhang WJ, Jia RZ (2012) Suppression of inflammation by tumor-derived exosomes: a kind of natural liposome packaged with multifunctional proteins. J Liposome Res 22:346–352
Liu GJ, Werry EL, Bennett MR (2005) Secretion of ATP from Schwann cells in response to uridine triphosphate. Eur J Neurosci 21:151–160. https://doi.org/10.1111/j.1460-9568.2004.03831.x
De-Xiang B, Guang-Zhi N, Shi-Qing F, Ying W, Xian-Hu Z, Yang L, Jia-Tong C (2011) Combination of activated Schwann cells with bone mesenchymal stem cells: the best cell strategy for repair after spinal cord injury in rats. Regen Med 6:707–720
Kaewkhaw R, Scutt AM, Haycock JW (2012) Integrated culture and purification of rat Schwann cells from freshly isolated adult tissue. Nat Protoc 7:1996–2004. https://doi.org/10.1038/nprot.2012.118
Chang YM, Chang HH, Tsai CC, Lin HJ, Ho TJ, Ye CX, Chiu PL, Chen YS, Chen RJ, Huang CY (2017) Alpinia oxyphylla Miq. fruit extract activates IGFR-PI3K/Akt signaling to induce Schwann cell proliferation and sciatic nerve regeneration. Bmc Complement Altern Med 17:184
Yuhua J, Mi S, Xin W, Shuqiang Z, Shu Y, Gang C, Xiaosong G, Fei D (2012) Comparative proteomic analysis of primary schwann cells and a spontaneously immortalized schwann cell line RSC 96: a comprehensive overview with a focus on cell adhesion and migration related proteins. J Proteome Res 11:3186–3198
Goldman SA, Maiken N, Windrem MS (2012) Glial progenitor cell-based treatment and modeling of neurological disease. Science 338:491–495
Court FA, Hendriks WTJ, Macgillavry HD, Jaime A, Jan VM (2008) Schwann cell to axon transfer of ribosomes: toward a novel understanding of the role of glia in the nervous system. J Neurosci 28:11024–11029
Stankiewicz TR, Linseman DA (2014) Rho family GTPases: key players in neuronal development, neuronal survival, and neurodegeneration. Frontiers Cell Neurosci 8:314
Matsukawa T, Morita K, Omizu S, Kato S, Koriyama Y (2018) Mechanisms of RhoA inactivation and CDC42 and Rac1 activation during zebrafish optic nerve regeneration. Neurochem Int 112:71–80. https://doi.org/10.1016/j.neuint.2017.11.004
Renault-Mihara F, Mukaino M, Shinozaki M, Kumamaru H, Kawase S, Baudoux M, Ishibashi T, Kawabata S, Nishiyama Y, Sugai K (2017) Regulation of RhoA by STAT3 coordinates glial scar formation. J Cell Biol 216:2533
Zeug A, Müller FE, Anders S, Herde MK, Minge D, Ponimaskin E, Henneberger C (2018) Control of astrocyte morphology by Rho GTPases. Brain Res Bull 136:44–53
An Y, Liu T, Liu X, Zhao L, Wang J (2016) Rac1 and Cdc42 play important roles in arsenic neurotoxicity in primary cultured rat cerebellar astrocytes. Biol Trace Elem Res 170:173–182. https://doi.org/10.1007/s12011-015-0456-7
Neubrand VE, Pedreno M, Caro M, Forte-Lago I, Delgado M, Gonzalez-Rey E (2014) Mesenchymal stem cells induce the ramification of microglia via the small RhoGTPases Cdc42 and Rac1. Glia 62:1932–1942. https://doi.org/10.1002/glia.22714
Yingying D, Yun G, Youjuan H, Yingjie W, Yan L, Mei L, Fei D, Xiaosong G, Yongjun W (2013) HMGB1 protein does not mediate the inflammatory response in spontaneous spinal cord regeneration: a hint for CNS regeneration. J Biol Chem 288:18204–18218
Zheng B, Ye L, Zhou Y, Zhu S, Wang Q, Shi H, Chen D, Wei X, Wang Z, Li X (2016) Epidermal growth factor attenuates blood-spinal cord barrier disruption via PI3K/Akt/Rac1 pathway after acute spinal cord injury. J Cell Mol Med 20:1062–1075
Acknowledgements
We thanked the support of the following funding: NSFC Program (81330042, 81620108018, 81672171, 81702147), Ministry of Science and Technology, China (2014DFR31210), Tianjin Science and Technology Committee, China (13RCGFSY19000, 14ZCZDSY00044).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
No competing interests exist in the submission of this manuscript.
Conflict of interest
All authors declare no conflict of interests and no disclosures relevant to the manuscript.
Ethical approval
All authors have read the Journal’s position on issues involved in ethical publication, and all authors have approved the final version of the manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wei, Z., Fan, B., Ding, H. et al. Proteomics analysis of Schwann cell-derived exosomes: a novel therapeutic strategy for central nervous system injury. Mol Cell Biochem 457, 51–59 (2019). https://doi.org/10.1007/s11010-019-03511-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-019-03511-0