Abstract
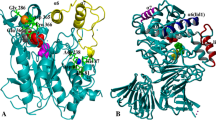
In the present investigation, we used directed evolution approach to engineer a lipase from metagenomic origin. A variant S311C, was generated, characterized in detail and compared with wild type. Wild type and variant lipases were overexpressed and purified to homogeneity. The temperature optima of the purified lipases (Variant and wild type) were almost same, and found to be 45 and 50 °C, respectively. The variant protein was highly thermostable (54 times) as compared with the wild type at 60 °C. The variant displayed very high kinetic efficiency over the wild type protein. Analysis of the homology models of wild type and variant lipase showed that the substitution is on the surface of the protein. This substitution, along with hydrophobic residues in near vicinity may be involved in formation of strong hydrophobic channel leading to active site. This study identifies the role of hydrophobic interactions in protein stability along with enhancement of enzyme activity.




Similar content being viewed by others
References
Kourist R, Brundiek H, Bornscheuer UT (2010) Protein engineering and discovery of lipases. Eur J Lipid Sci Technol 112:64–74
Anthonsen HW, Baptista A, Drablos F, Martel P, Petersen SB, Sebastiao M, Vaz L (1995) Lipases and esterases: a review of their sequences, structure and evolution. Biotechnol Annu Rev 1:315–371
Verger R (1997) ‘Interfacial activation’ of lipases: facts and artefacts. Trends Biotechnol 15:32–38
Schmid RD, Verger R (1998) Lipases—interfacial enzymes with attractive applications. Angew Chem Int Ed 37:1608–1633
Noel M, Combes D (2003) Rhizomucor miehei lipase: differential scanning calorimetry and pressure/temperature stability studies in presence of soluble additives. Enzyme Microb Technol 33:299–308
Weber N, Klein E, Vosmann K, Mukherjee KD (2004) Mono-thioesters and di-thioesters by lipase-catalyzed reactions of alpha, omega alkanedithiols with palmitic acid or its methyl ester. Appl Microbiol Biotechnol 64:800–805
Weber N, Bergander K, Fehling E, Klein E, Vosmann K, Mukherjee KD (2006) Copolymeric polythioesters by lipase-catalyzed thioesterification and transthioesterification of alpha, omega-alkanedithiols. Appl Microbiol Biotechnol 70:290–297
Keng PS, Basri M, Ariff AB, Abdul Rahman MB, Abdul Rahman RN, Salleh AB (2008) Scale-up synthesis of lipase-catalyzed palm esters in stirred-tank reactor. Bioresour Technol 99:6097–6104
Peterson ME, Daniel RM, Danson MJ, Eisenthal R (2007) The dependence of enzyme activity with temperature: determination and validation of parameters. Biochem J. 402:331–337
Romero PA, Arnold FH (2009) Exploring protein fitness landscapes by directed evolution. Nature Review Mol Cell Biol 10:866–876
Mills DR, Peterson RL, Spiegelman S (1967) Proc Natl Acad Sci USA 58:217–224
Reetz MT (2004) Controlling the enantioselectivity of enzymes by directed evolution: practical and theoretical ramifications. Proc Natl Acad Sci USA 101:5716–5722
May O, Nguyen PT, Arnold FH (2000) Inverting enantioselectivity by directed evolution of hydantoinase for improved production of l-methionine. Nat Biotechnol 18(3):317–320
Choi WC, Kim MH, Ro HS, Sang RR, Oh TK, Lee JK (2005) Zinc in lipase L1 from Geobacillus stearothermophilus L1 and structural implications on thermal stability. FEBS Lett 579:3461–3466
Zhou J, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322
Sharma PK, Capalsh N, Kaur J (2007) An improved method for single step purification of metagenomic DNA. Mol Biotechnol 36:61–63
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291
Luthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356:83–85
Wiederstein, Sippl (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:W407–W410
CCP4 Newsletter (Number 42, Summer 2005)
Nawani N, Kaur J (2000) Purification, characterization and thermostability of lipase from a thermophilic Bacillus sp. Mol Cell Biochem 206:91–96
Saxena RK, Sheoran A, Giri B, Davidson WS (2009) Purification strategies for microbial lipases. J Microbiol Methods 52:1–18
Pastore GM, Costa VSR, Koblitz MGB (2003) Partial purification and biochemistry characterization of an extracellular lipase obtained from new Rhizopus sp. stream. Ciência e Tecnologia de Alimentos 23(2):135–140
Pencreac’h G, Barrati JC (1996) Hydrolysis of p-nitrophenyl palmitate in n-heptane by the Pseudomonas cepacia lipase: a simple test for the determination of lipase activity in organic media. Enzym Microb Technol 18:417–422
Sharma PK, Singh K, Singh R, Capalash N, Ali A, Mohammad O, Kaur J (2012) Characterization of thermostable lipase showing loss of secondary structure at ambient temperature. Mol Biol Rep 39(3):2795–2804
Sharma PK, Kumar R, Kumar R, Mohammad O, Singh R, Kaur J (2012) Engineering of a metagenome derived lipase toward thermal tolerance: effect of asparagine to lysine mutation on the protein surface. Gene 491(2):264–271
Zhao H, Arnold FH (1999) Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Eng 12(1):47–53
Khurana J, Singh R, Kaur J (2010) Engineeering of Bacillus lipase by directed evolution for enhanced thermal stability: effect of isoleucine to threonine mutation. Mol Biol Rep 38:2919–2926
Castro RQ, Diaz P, Alfaro GV, Gracia HS, Ros RO (2009) Gene cloning, expression and characterization of the Geobacillus thermoleovorans CCR11 thermoalkaliphilic Lipase. Mol Biotechnol 42:75–83
Kim HK, Park SY, Lee JK, Oh TK (1998) Gene cloning and characterization of thermostable lipase from Bacillus stearothermophilus L1. Biosci Biotechnol Biochem 62:66–71
Sinchaikul S, Sookkheo B, Phutrakul S, Pan FM, Chen ST (2001) Optimization of a thermostable lipase from Bacillus stearothermophilus P1; overexpression, purification, and characterization. Protein Expr Purif 22:388–398
Johannes TW, Woodyer RD, Zhao H (2005) Directed evolution of a thermostable phosphite dehydrogenase for NAD(P)H regeneration. Appl Environ Microbiol 71:5734–5756
Soliman NA, Knoll M, Fattah YRA, Schmid RD, Lange S (2007) Molecular cloning and characterization of thermostable esterase and lipase from Geobacillus thermoleovorans YN isolated from desert soil in Egypt. Process Biochem 42:1090–1100
Li H, Zhang X (2005) Characterization of thermostable lipase from thermophilllic Geobacillus sp. Protein Expr Purif 42:153–159
Nakatani T, Hiratake J, Yoshikawa K, Nishioka T, Oda J (1992) Chemical inactivation of lipase in organic solvent: a lipase from Pseudomonas aeruginosa TE3285 is more like a typical serine enzyme in an organic solvent than in aqueous media. Biosci Biotechnol Biochem 56:1118–1123
Dannert CS, Sztajer H, Stocklein W, Menge U, Schmid RD (1994) Screening, purification and properties of a thermophilic lipase from Bacillus thermocatenulatus. Biochim Biophys Acta 1214:43–53
Reetz M, Soni P, Ferna′ndez L, Gumulya Y, Carballeira J (2010) Increasing the stability of an enzyme toward hostile organic solvents by directed evolution based on iterative saturation mutagenesis using the B-FIT method. Chem Commun 46:8657–8658
Jeong ST, Kim HK, Kim SJ, Chi SW, Pan JG, Oh TK, Ryu SE (2002) Novel Zinc-binding Center and a Temperature Switch in the Bacillus stearothermophilus L1 Lipase. J Biol Chem 277:17041–17047
Acknowledgments
Author acknowledges the financial assistance to Dr. Jagdeep Kaur from the Department of Science and Technology, New Delhi, India.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ranvir Singh and Jagdeep Kaur contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11010_2012_1483_MOESM2_ESM.doc
SD Fig-1: Showing multiple sequence alignment of Wild type (pJKR1), Uncultured bacterium (JKP01), BSL, BSP and SF1 having accession no- AEL99899, ACJ07039, AAC12257, AAF40217 and AAY82869 respectively. Active site residues have been shown in blue colours (Ser is in conserved pentapeptide (AHSQG)). Signal peptide is shown is yellow colour. (DOC 32 kb)
11010_2012_1483_MOESM3_ESM.doc
SD Fig-2: Hydropathy plots of the proteins LipR1 (5A) and LipR2 (5B). Blue lines denote Hydropathy. Red lines denote Amphipathicity. The circles are showing region of change in hydropathy due to change in amino acid S311C. (DOC 230 kb)
Rights and permissions
About this article
Cite this article
Kumar, R., Sharma, M., Singh, R. et al. Characterization and evolution of a metagenome-derived lipase towards enhanced enzyme activity and thermostability. Mol Cell Biochem 373, 149–159 (2013). https://doi.org/10.1007/s11010-012-1483-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-012-1483-8