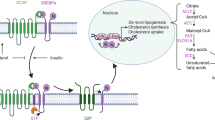
Abstract
Bile acids subserve important physiological functions in the control of cholesterol homeostasis. Indeed, hepatic bile acid synthesis and biliary excretion constitute the main route for cholesterol removal from the human body. On the other hand, bile acids serve as natural detergents for the intestinal absorption of dietary cholesterol. However, due to their detergent properties, bile acids are inherently cytotoxic, and their cellular level may be tightly controlled to avoid pathological situations such as cholestasis. Recent investigations have illustrated the crucial roles that a series of ligand-activated transcription factors has in the control of hepatic bile acids synthesis, transport and metabolism. Thus, the lipid-activated nuclear receptors, farnesoid X-receptor (FXR), liver X-receptor (LXR), pregnane X-receptor (PXR) and peroxisome proliferator-activated receptor alpha (PPARα), modulate the expression and activity of genes controlling bile acid homeostasis in the liver. Several members of the UDP-glucuronosyltransferase (UGT) enzymes family are among the bile acid metabolizing enzymes regulated by these receptors. UGTs catalyze glucuronidation, a major phase II metabolic reaction, which converts hydrophobic bile acids into polar and urinary excretable metabolites. This article summarizes our recent observations on the regulation of bile acid conjugating UGTs upon pharmacological activation of lipid-activated receptors, with a particular interest for the role of PPARα and LXRα in controlling human UGT1A3 expression.


Similar content being viewed by others
References
Russell DW (2003) The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem 72:137–174. doi:10.1146/annurev.biochem.72.121801.161712
Chiang JY (2002) Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocr Rev 23:443–463. doi:10.1210/er.2000-0035
Trottier J, Milkiewicz P, Kaeding J et al (2006) Coordinate regulation of hepatic bile acid oxidation and conjugation by nuclear receptors. Mol Pharm 3:212–222. doi:10.1021/mp060020t
Bodin K, Lindbom U, Diczfalusy U (2005) Novel pathways of bile acid metabolism involving CYP3A4. Biochim Biophys Acta 1687:84–93. doi:10.1016/j.bbalip.2004.11.003
Pauli-Magnus C, Meier PJ (2005) Hepatocellular transporters and cholestasis. J Clin Gastroenterol 39:S103–S110. doi:10.1097/01.mcg.0000155550.29643.7b
Marschall HU, Matern H, Wietholtz H et al (1992) Bile acid N-acetylglucosaminidation: in vivo and in vitro evidence for a selective conjugation reaction of 7 beta-hydroxylated bile acids in humans. J Clin Invest 89:1981–1987. doi:10.1172/JCI115806
Takikawa H, Otsuka H, Beppu T et al (1983) Serum concentrations of bile acid glucuronides in hepatobiliary diseases. Digestion 27:189–195
Barbier O, Bélanger A (2003) The cynomolgus monkey (Macaca fascicularis) is the best animal model for the study of steroid glucuronidation. J Steroid Biochem Mol Biol 85:235–245. doi:10.1016/S0960-0760(03)00235-8
Bélanger A, Pelletier G, Labrie F et al (2003) Inactivation of androgens by UDP-glucuronosyltransferase enzymes in humans. Trends Endocrinol Metab 14:473–479. doi:10.1016/j.tem.2003.10.005
Mackenzie PI, Walter Bock K, Burchell B et al (2005) Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics 15:677–685. doi:10.1097/01.fpc.0000173483.13689.56
Alme B, Sjovall J (1980) Analysis of bile acid glucuronides in urine: identification of 3 alpha, 6 alpha, 12 alpha-trihydroxy-5 beta-cholanoic acid. J Steroid Biochem 13:907–916. doi:10.1016/0022-4731(80)90164-8
Meng LJ, Reyes H, Palma J et al (1997) Profiles of bile acids and progesterone metabolites in the urine and serum of women with intrahepatic cholestasis of pregnancy. J Hepatol 27:346–357. doi:10.1016/S0168-8278(97)80181-X
Lévesque E, Beaulieu M, Hum DW et al (1999) Characterization and substrate specificity of UGT2B4 (E458): a UDP-glucuronosyltransferase encoded by a polymorphic gene. Pharmacogenetics 9:207–216
Pillot T, Ouzzine M, Fournel-Gigleux S et al (1993) Glucuronidation of hyodeoxycholic acid in human liver: evidence for a selective role of UDP-glucuronosyltransferase 2B4. J Biol Chem 268:25636–25642
Gall WE, Zawada G, Mojarrabi B et al (1999) Differential glucuronidation of bile acids, androgens and estrogens by human UGT1A3 and 2B7. J Steroid Biochem Mol Biol 70:101–108. doi:10.1016/S0960-0760(99)00088-6
Back P (1976) Bile acid glucuronides, II[1]: isolation and identification of a chenodeoxycholic acid glucuronide from human plasma in intrahepatic cholestasis. Hoppe Seylers Z Physiol Chem 357:213–217
Ikegawa S, Okuyama H, Oohashi J et al (1999) Separation and detection of bile acid 24-Glucuronides in human urine by liquid chromatography combined electrospray ionization mass spectrometry. Anal Sci 15:625–631. doi:10.2116/analsci.15.625
Takikawa H, Beppu T, Seyama Y (1985) Serum concentrations of glucuronidated and sulfated bile acids in children with cholestasis. Biochem Med 33:381–386. doi:10.1016/0006-2944(85)90014-6
Trottier J, Verreault M, Grepper S et al (2006) Human UDP-glucuronosyltransferase (UGT)1A3 enzyme conjugates chenodeoxycholic in the liver. Hepatology 44:1158–1170. doi:10.1002/hep.21362
Caron P, Trottier J, Verreault M et al (2006) Enzymatic production of bile acid glucuronides used as analytical standards for liquid chromatography-mass spectrometry analyses. Mol Pharm 3:293–302. doi:10.1021/mp060021l
Johnson DW (2005) Contemporary clinical usage of LC/MS: analysis of biologically important carboxylic acids. Clin Biochem 38:351–361. doi:10.1016/j.clinbiochem.2005.01.007
Eloranta JJ, Kullak-Ublick GA (2005) Coordinate transcriptional regulation of bile acid homeostasis and drug metabolism. Arch Biochem Biophys 433:397–412. doi:10.1016/j.abb.2004.09.019
Verreault M, Senekeo-Effenberger K, Trottier J et al (2006) The liver X-receptor alpha controls hepatic expression of the human bile acid-glucuronidating UGT1A3 enzyme in human cells and transgenic mice. Hepatology 44:368–378. doi:10.1002/hep.21259
Senekeo-Effenberger K, Chen S, Brace-Sinnokrak E et al (2007) Expression of the human UGT1 locus in transgenic mice by 4-chloro-6-(2,3-xylidino)-2-pyrimidinylthioacetic acid (WY-14643) and implications on drug metabolism through peroxisome proliferator-activated receptor {alpha} activation. Drug Metab Dispos 35:419–427. doi:10.1124/dmd.106.013243
Barbier O, Torra IP, Duguay Y et al (2002) Pleiotropic actions of peroxisome proliferator-activated receptors in lipid metabolism and atherosclerosis. Arterioscler Thromb Vasc Biol 22:717–726. doi:10.1161/01.ATV.0000015598.86369.04
Sonoda J, Pei L, Evans RM (2008) Nuclear receptors: decoding metabolic disease. FEBS Lett 582:2–9. doi:10.1016/j.febslet.2007.11.016
Post SM, Duez H, Gervois PP et al (2001) Fibrates suppress bile acid synthesis via peroxisome proliferator-activated receptor-alpha-mediated downregulation of cholesterol 7alpha-hydroxylase and sterol 27-hydroxylase expression. Arterioscler Thromb Vasc Biol 21:1840–1845. doi:10.1161/hq1101.098228
Hunt MC, Yang YZ, Eggertsen G et al (2000) The peroxisome proliferator-activated receptor alpha (PPARalpha) regulates bile acid biosynthesis. J Biol Chem 275:28947–28953. doi:10.1074/jbc.M002782200
Zimniak P, Holsztynska EJ, Lester R et al (1989) Detoxification of lithocholic acid. Elucidation of the pathways of oxidative metabolism in rat liver microsomes. J Lipid Res 30:907–918
Repa JJ, Mangelsdorf DJ (2000) The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Annu Rev Cell Dev Biol 16:459–481. doi:10.1146/annurev.cellbio.16.1.459
Barbier O, Duran-Sandoval D, Pineda-Torra I et al (2003) Peroxisome proliferator-activated receptor alpha induces hepatic expression of the human bile acid glucuronidating UDP-glucuronosyltransferase 2B4 enzyme. J Biol Chem 278:32852–32860. doi:10.1074/jbc.M305361200
Acknowledgments
Most of the data summarized here would not have been obtained without the efficient contribution of our collaborators: members of Prof. Chantal Guillemette’s and Alain Bélanger’s laboratories (Genomic and Endocrinology Research Center, CHUQ Research Center, Laval university, Québec, Canada), of Prof. Robert H. Tukey’s lab (Laboratory of Environmental Toxicology, Department of Chemistry & Biochemistry and Department of Pharmacology, University of California, San Diego, La Jolla, CA, USA) and of Prof. Bart Staels’ group (Unité 545, INSERM, Lille and Department of Atherosclerose, Institute Pasteur de Lille and the Faculté de Pharmacie, Université de Lille II, Lille, F-59019, France). We thank Dr. Virginie Bocher for critical reading of this manuscript. Financial support: This work was supported by the “Canadian Institutes for Health Research CIHR” (MOP 118446), the “Fonds pour la Recherche en Santé du Québec; FRSQ”. Olivier Barbier is supported by the Health Research Foundation of Rx&D-CIHR. Jenny Kaeding is holder of a scholarship from the Health Research Foundation of Rx&D-CIHR. Jocelyn Trottier is holder of a scholarship from CIHR.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Barbier, O., Trottier, J., Kaeding, J. et al. Lipid-activated transcription factors control bile acid glucuronidation. Mol Cell Biochem 326, 3–8 (2009). https://doi.org/10.1007/s11010-008-0001-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-008-0001-5