Abstract
Hepatocellular carcinoma (HCC) is an increasing burden on global public health and is associated with enhanced lipogenesis, fatty acid uptake, and lipid metabolic reprogramming. De novo lipogenesis is under the control of the transcription factor sterol regulatory element-binding protein 1 (SREBP-1) and essentially contributes to HCC progression. Here, we summarize the current knowledge on the regulation of SREBP-1 isoforms in HCC based on cellular, animal, and clinical data. Specifically, we (i) address the overarching mechanisms for regulating SREBP-1 transcription, proteolytic processing, nuclear stability, and transactivation and (ii) critically discuss their impact on HCC, taking into account (iii) insights from pharmacological approaches. Emphasis is placed on cross-talk with the phosphatidylinositol-3-kinase (PI3K)-protein kinase B (Akt)-mechanistic target of rapamycin (mTOR) axis, AMP-activated protein kinase (AMPK), protein kinase A (PKA), and other kinases that directly phosphorylate SREBP-1; transcription factors, such as liver X receptor (LXR), peroxisome proliferator-activated receptors (PPARs), proliferator-activated receptor γ co-activator 1 (PGC-1), signal transducers and activators of transcription (STATs), and Myc; epigenetic mechanisms; post-translational modifications of SREBP-1; and SREBP-1-regulatory metabolites such as oxysterols and polyunsaturated fatty acids. By carefully scrutinizing the role of SREBP-1 in HCC development, progression, metastasis, and therapy resistance, we shed light on the potential of SREBP-1-targeting strategies in HCC prevention and treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver cancer, accounting for approximately 90% of cases, and is often accompanied by hepatitis B virus (HBV) infection or non-alcoholic steatohepatitis (NASH) as leading risk factors [1, 2]. Malignant transformation to HCC is associated with intense metabolic reprogramming, particularly related to lipid uptake, biosynthesis, (subcellular) transport, distribution, degradation, and signaling [3]. Compared to non-tumorous liver cells, which largely acquire fatty acids from extracellular sources, HCC cells develop a remarkable ability to synthesize lipids de novo while upregulating fatty acid uptake [4, 5]. Thus, HCC initiation and progression are highly dependent on lipogenic enzymes, such as fatty acid synthase (FASN), stearoyl-coenzyme A desaturase (SCD), acetyl-coenzyme A carboxylase 1 (ACC1), and malic enzyme (ME), and upstream regulators, including sterol regulatory element-binding protein 1 (SREBP-1), all of which are highly expressed in human HCC [4,5,6]. As a central lipid-anabolic transcription factor, SREBP-1 regulates a variety of (rate-limiting) enzymes in fatty acid and triglyceride biosynthesis, including ATP-citrate lyase (ACLY), ACC1/2, FASN, and SCD1 [7]. High expression of SREBP-1 and its target enzymes predicts poor survival of HCC patients and is associated with increased tumor size, high histological grade, and advanced tumor-node-metastasis stage [8, 9]. In summary, SREBP-1 plays an important role in HCC tumorigenesis and metastasis [7, 10, 11], serves as an independent prognostic marker for overall and disease-free survival of HCC patients [9], and represents a promising target for therapeutic intervention [12, 13].
2 SREBP-1
SREBP-1 belongs to the SREBP family of transcription factors and contains a basic helix-loop-helix-leucine zipper (bHLH-Zip) domain for binding to sterol regulatory elements (SREs) (5′-ATCACCCCAC-3′) and E-boxes (5′-CANNTG-3′; sterol-independent SREBP interaction sites) on target gene promoters [10, 14]. SREs are also located at SREBP promoters, and autoregulatory loops have been described [15]. The three members, SPREB-1a, SREBP-1c, and SREBP-2, are encoded by two genes, SREBF1 (for SREBP-1a and c) and SREBF2 (for SREBP-2) [10]. SREBP-1a and SREBP-1c are encoded by a single gene and are transcribed from alternative start sites [16]. Accordingly, the N-terminal sequence of SREBP-1c is shorter than that of SREBP-1a, resulting in weaker (hepatic) transcriptional activity [17, 18]. Germline deletion of Srebp-2 results in 100% lethality in mice, while the majority of homozygous Srebp-1-knockout mice do not survive in utero, primarily due to the loss of Srebp-1a [16]. SREBP-1a is a potent inducer of SREBP-responsive genes, regardless of whether they are involved in cholesterol, fatty acid, or triglyceride metabolism [16]. The functions of SREBP-1c are more restricted and concentrate on the expression of genes required for fatty acid biosynthesis [16]. While SREBP-2 is widely expressed [10], SREBP-1c and SREBP-1a show distinct tissue specificity: SREBP-1c is the predominant isoform in hepatocytes, and SREBP-1a is highly expressed in adipocytes and certain hepatomas [17].
SREBPs are synthesized as inactive SREBP precursors (pSREBPs), which are inserted into the endoplasmic reticulum (ER) membrane and bind to the COOH-terminal region of SREBP cleavage-activating protein (SCAP) (Fig. 1) [10, 19]. The N-terminal region of SCAP functions as a sterol-sensing domain. At high cellular sterol levels, SCAP binds cholesterol, undergoes a conformational change, and in complex with SREBP interacts with insulin-induced gene 1 proteins (Insig1 and Insig2) [19]. These ER-resident membrane proteins bind cholesterol, block ER exit, and prevent the subsequent transfer to the Golgi [19]. When cellular sterol levels drop, Insig1 is ubiquitylated by the E3 ubiquitin ligase gp78 and undergoes proteasomal degradation [20, 21]. SCAP-SREBP dissociates from Insigs and is transferred to the Golgi, where SREBPs are subjected to proteolytic cleavage by site-1 protease (S1P) and subsequently S2P [22]. The mature N-terminal SREBP bHLH-Zip domain (mSREBP) is released into the cytosol, enters the nucleus as a homodimer, and binds to SRE- or E-box-containing promoters [13]. Cholesterol-derived oxysterols (e.g., 25-hydroxycholesterol) indicate excess cholesterol and mediate ER retention of the SCAP-SREBP complex by binding to Insigs [23, 24].
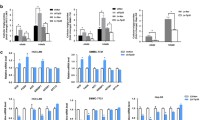
Proteolytic processing of SREBP-1 stimulates de novo lipogenesis and sustains tumorigenesis. pSREBP binds to SCAP and is anchored to the ER membrane by Insigs. Under low cellular sterol conditions, the SCAP-SREBP complex is released from Insigs and transferred to the Golgi, where pSREBP is cleaved by S1P and S2P, releasing the soluble mSREBP into the cytosol. mSREBP forms homodimers and translocates to the nucleus, binds to promoters with SREs or E-boxes, and transactivates the transcription of target genes. By inducing the expression of lipogenic enzymes (ACLY, ACC, FASN, elongation of long fatty acids family member 6 (ELOVL6), and SCD shown in purple) and enhancing de novo lipogenesis, SREBP-1 promotes membrane biogenesis, which is essential for tumor growth. Figure modified from Horton et al. and Moon et al. [16, 25] and created with https://www.biorender.com/
3 Role of SREBP-1 in HCC progression
Lipids serve as energy reservoirs, structural components, and signaling mediators vital for maintaining cell growth and proliferation [11]. Research has highlighted the importance of lipid metabolism in driving cancer phenotypes [7], adapting to stress responses, including lipotoxicity [26], regulating cell survival [7, 27], and shaping the interplay of cancer cells with immunity and the tumor microenvironment [28,29,30,31], thereby revealing potential targets for anti-cancer therapy [32, 33]. This multifaceted landscape is tightly controlled by a multitude of enzymes and other factors responsible for lipid biosynthesis, lipid catabolism, and energy balance, which are subject to rigorous transcriptional and post-transcriptional regulation [11].
The transcription factor SREBP-1 serves as a central driver of lipogenesis, regulating the expression of multiple lipogenic enzymes involved in the biosynthesis of cholesterol, fatty acids, and triglycerides. It acts as a hub for myriad physiological and pathophysiological cellular processes, functioning in both transcriptional and post-transcriptional regulation [16, 34]. SREBP-1 promotes cell growth and viability, and loss of SREBP-1 signaling leads to severe lipotoxicity in glioma cells [26]. This lipotoxicity can be mitigated by the addition of monounsaturated fatty acids (MUFAs) and is considered to result from an imbalance between saturated fatty acids and MUFAs due to disturbed desaturation by SCD1 [26]. SREBP-1 is also involved in the regulation of ER stress and cell death. In osteosarcoma cells, the overexpression of SREBP-1 inhibits cell proliferation, upregulates the expression and phosphorylation of protein kinase RNA-like endoplasmic reticulum kinase (PERK), and amplifies the PERK-activated unfolded protein response (UPR), leading to ER stress-induced apoptosis and autophagy [35]. Silencing of SREBP-1 attenuates this stress response [35], whereas SREBP-1 upregulation by high glucose promotes cell growth and inhibits apoptosis and autophagy in pancreatic cancer cells [36]. Furthermore, SREBP-1 decreases ferroptosis sensitivity by inducing the SCD1-MUFA axis and modulates inflammatory reactions, such as those induced by obesity and toll-like receptor 4 (TLR4) [37,38,39].
The regulation of SREBP-1 is at the core of numerous metabolic diseases, such as obesity, diabetes, atherosclerosis, non-alcoholic fatty liver disease, hepatosteatosis, neurodegenerative diseases, and cancer [40,41,42,43,44]. The latter relies on membrane biogenesis and energy production for cell growth and proliferation (Fig. 1) and is often associated with enhanced SREBP-1 expression and/or activation [45,46,47]. In HCC, SREBP-1 activates a comprehensive lipogenic program [9], which includes the transcriptional upregulation of FASN, ACC, SCD, and ACLY and downregulation of medium-chain acyl-CoA dehydrogenase (ACADM)–mediated fatty acid oxidation [48]. SREBP-1 has been implicated in (i) promoting HCC proliferation and survival [13, 49,50,51,52,53,54,55,56,57,58,59], with high levels of SREBP-1 correlating with increased mortality in HCC patients [60]. Accordingly, the recently discovered SREBP-1 inhibitor, cinobufotalin, dose-dependently reduced both the HCC tumor volume and weight in tumor-bearing mice after 2 weeks of treatment [61]. In diethylnitrosamine (DEN)-induced HCC in C57 mice, dietary cholesterol suppressed HCC progression, which was ascribed to the inhibition of SCAP-dependent de novo lipogenesis [55]. Cholesterol deprivation had the opposite effect. (ii) SREBP-1 prevents cell death induction by adjusting lipid composition and content, thereby reducing the susceptibility of cell membranes to peroxidation and alleviating cellular lipotoxicity [51, 53, 62, 63]. (iii) SREBP-1 also increased tumor stemness, possibly by promoting ACLY-induced metabolic plasticity [13, 45, 64], and (iv) triggered epithelial-mesenchymal transition (EMT), migration, and metastasis, which is related to the regulation of Drosophila embryonic protein (Snail) stability and enhanced glycolytic activity [9, 13, 46, 52, 65, 66]. Thus, HCC knockdown prominently inhibited cell migration and invasion in HCC cell lines [9]. (v) SREBP-1 promotes vascular endothelial growth factor (VEGF)–induced angiogenesis [46], (vi) contributes to inflammation sensing and the generation of a tumor-promoting, pro-inflammatory microenvironment [29, 38, 39, 53, 67, 68], which may involve a feed-forward loop in which androgen and interleukin 6 (IL-6)–activated cell cycle–related kinase (CCRK) promotes non-alcoholic steatohepatitis (NASH)–derived HCC by activating the mTOR-SREBP-1 axis [69]. Blocking the SREBP pathway by liver-specific ablation of gp78 or SCAP suppressed DEN-induced hepatocarcinogenesis, resulting in reduced levels of tumor-promoting cytokines, including IL-6, IL-1β, and tumor necrosis factor-alpha (TNF-α), in mice [38]. In addition, dendritic cells exposed to HCC-derived α-fetoprotein decreased SREBP-1 expression and fatty acid biosynthesis, leading to immune suppression [70]. (vii) SREBP-1 increased HCC therapy resistance [27, 37, 44, 66, 71,72,73,74], including SCD1- or glycolytic-activated chemoresistance [37, 66], radioresistance through a combination of glucose and cardiolipin anabolism [71], and transforming growth factor beta 1 (TGF-β1)–enhanced immunoresistance [73]. Accordingly, inhibition of SREBP-1 activity by small-molecule inhibitors such as SI-1 or betulin increased the sensitivity of HCC tissue to radiofrequency ablation or sorafenib, respectively [44, 66].
The functions of SREBP-1 have been extensively reviewed, and the mechanisms behind SREBP-1 regulation, as well as its link to tumorigenesis, metabolism, metastasis, immune evasion, and therapy resistance, have been thoroughly addressed in recent reviews [11,12,13, 16, 28, 53, 75, 76]. While many of these articles refer to HCC and/or pre-malignant states, systematic reviews on HCC have been lacking. However, the emerging understanding of cancer heterogeneity demands an organ-, tissue-, cell type-, and context-dependent view on putative therapeutic strategies. In the following, we summarize and critically discuss the current knowledge on the regulation of SREBP-1 in HCC and highlight pharmacological approaches directed against HCC for which interference with SREBP-1 signaling has been confirmed.
4 Diversity of factors controlling SREBP-1 signaling in HCC
SREBP-1 signaling is regulated in HCC by a variety of factors, including (i) endocrine hormones such as insulin [18, 77,78,79,80,81], glucagon [80, 82, 83], thyrotropin (TSH) [84], thyroid hormones [85, 86], glucocorticoids [87], and sex hormones (dihydrotestosterone) [88]; (ii) para- and autocrine peptide hormones such as growth factors (hepatocyte growth hormone (HGF), hepatoma-derived growth factor (HDGF), fibroblast growth factor 21 (FGF21)) [87, 89,90,91], cytokines (TNF-α, IL-6, IL-17A, TGFβ) [69, 92,93,94,95,96], and interferon-γ (IFN-γ) [73]; (iii) membrane-localized receptor ligands such as ephrin A3 [45]; (iv) lipid mediators, e.g., prostaglandin E2 (PGE2) [57]; (v) metabolites such as (oxy)sterols [23], bile acids [97], amino acids [98], and polyunsaturated fatty acids (PUFAs) [99,100,101]; and (vi) hepatitis viruses such as HBV and HCV [60], to name a few. Many of these factors use common signaling routes to regulate SREBP-1 signaling. For example, insulin and growth factors signal largely through RTKs, activating the phosphatidylinositol-3-kinase (PI3K)-protein kinase B (Akt)-mechanistic target of rapamycin (mTOR) axis [102], whereas the mechanistically more diverse cytokine receptors trigger different SREBP-1-regulating signaling cascades, including STAT signaling via transmembrane receptors with associated tyrosine kinases [103, 104]. Glucagon, many lipid mediators (including PGE2), and some cytokines/interferons activate or inhibit adenylate cyclase via G-protein-coupled 7-transmembrane receptors (GPCRs), thereby controlling PKA activation through cyclic 3′,5′-cyclic AMP (cAMP) generation, while having numerous cross-links to other pathways regulating SREBP-1 signaling [105,106,107]. Oxysterols, PUFAs, thyroid hormones, and bile acids bind to nuclear receptors (such as liver X receptor (LXR), farnesoid X receptor (FXR), and peroxisome proliferator-activated receptors (PPARs)) [108,109,110], and amino acids are an important element of nutrient sensing via mTOR [111]. Many additional endogenous factors belonging to these subgroups or sharing signaling pathways have not been explicitly reported to regulate SREBP-1 signaling in HCC, but have receptors on hepatocytes or other liver cells that regulate liver (lipid) metabolism and very likely also affect SREBP-1 signaling. Therefore, to structure the mechanisms in the control of SREBP-1 signaling, we have extracted the common pathways from the vast number of factors regulating SREBP-1 signaling in HCC and organized the following chapters accordingly.
5 Common pathways regulating SREBP-1 signaling in HCC
The activity of SREBP-1 is tightly regulated by a complex network of organ/tissue-specific signaling pathways at the level of (i) transcription; (ii) proteolytic processing and trafficking between the ER, Golgi, and nucleus; (iii) protein stability and degradation; and (iv) transactivating activity. It is generally accepted that SREBP-1a contributes to global lipid synthesis and growth [56, 112], whereas SREBP-1c represents a major insulin-responsive transcription factor that upregulates the expression of a variety of genes involved in (hepatic) fatty acid and triglyceride biosynthesis as well as glycolysis (i.e., glucokinase) [10, 66]. While this distinction is generally accurate, there are numerous exceptions and regulatory cross-talks, emphasizing the need for an organ/tissue/cell type–dependent understanding of the regulatory events. To provide a basis for the development of SREBP-1-targeting strategies specifically against HCC, we have compiled the current knowledge about the regulation of SREBP-1 in this cancer type (Fig. 2). We consider the tightly regulated signaling network controlling SREBP-1 transcription, maturation, subcellular localization, and transcriptional activity to be key to successful therapeutic approaches [32, 39, 50, 76]. It is now the time to address these challenges, given the recent mechanistic advances in understanding SREBP-1 regulation and signaling made possible by methodological breakthroughs in multiomics technologies, systems biology, genetic manipulation, and targeted (personalized) therapy.
Regulation of SREBP-1 in HCC. Focus is placed on the canonical pathways that link major regulators of SREBP-1 signaling to either SREBP-1 transcription, SREBP-1 mRNA stability, pSREBP-1 maturation and intracellular trafficking, mSREBP-1 post-translational modification affecting protein stability, degradation, and transactivating activity. Major regulatory pathways that coordinate the SREBP-1 signaling include (i) the PI3K-Akt-mTOR axis, thereby considering links to lipin-1, CRTC2, TIP30, and other PI3K-Akt-regulated pathways; (ii) the serine kinases AMPK and PKA as well as other kinases that directly phosphorylate SREBP-1; (iii) the transcription factors LXR, PPARs, STAT, Myc, and p53; (iv) histone acetyltransferases, sirtuins, PRMT5, and other factors that control post-translational modification of SREBP-1; (v) metabolites, including PUFAs; and (vi) various other regulatory factors and mechanisms, such as the microRNA miR-27a, the RNA binding proteins Lin28A and Lin28B, and nuclear-localized HDGF. Created with https://www.biorender.com/
Here, we concentrate on the different strategies how SREBP-1 is regulated during HCC initiation, progression, and metastasis as well as hepatic pathologies that contribute to HCC development and progression. Note that the latter studies have not explicitly investigated the impact on HCC. Furthermore, we highlight pharmacological approaches targeting liver cancer and for which a functional role of SREBP-1 interference has been confirmed or at least rationally proposed. This review is intended to help biomedical researchers working on HCC, as well as others with relevant expertise in cell metabolism, signal transduction, and lipid biology, to effectively use their interdisciplinary knowledge to address the remaining questions and find new strategies to combat aggressive (therapy-resistant) and recurrent HCC. Therefore, we comprehensively summarize the recent advances in SREBP-1 regulation; critically evaluate the conclusions drawn from the experimental data; highlight links to emerging fields, such as ferroptosis [42, 113]; and point out promising pharmacological strategies. The focus on SREBP-1, HCC, and related liver pathologies is of great importance in weighing the pros and cons of therapeutic approaches due to the substantial differences in SREBP-1 regulation between tissues, cell types, and metabolic states and distinguishes our review from others in the field. Another unique feature is that we put the spotlight on the interface between molecular signaling mechanisms, pharmacological targets, and therapeutic approaches. SREBP-1 target gene profiles and pro-tumoral signaling cascades are not the focus of this survey, and we would like to refer to excellent overview articles addressing these aspects [13, 39, 76, 114]. In addition, we would like to highlight a very recent study linking mTOR complex 1 (mTORC1)/SREBP-1c-dependent cardiolipin biosynthesis to cell survival and radioresistance in HCC [71]. Small-molecules targeting SREBP-1 signaling are listed in Table 1. Note that we only included articles that functionally linked SREBP-1 regulation/modulation to HCC or related liver pathologies and ignored others that described the effects of specific pathways or small molecules on HCC or SREBP-1 in independent studies and different systems [39, 76].
6 PI3K-AKT-mTOR axis
The PI3K-Akt-mTOR axis is a central signaling cascade initiated by receptor tyrosine kinases (RTKs), including the insulin receptor (IR) and growth factor receptors, and essentially regulates fatty acid, phospholipid, and neutral lipid metabolism by activating SREBP-1 [50, 195, 196]. There are manifold other pathways that converge in the PI3K-Akt-mTOR-SREBP-1-axis, including hypoxia-initiated signaling routes [57], the surface glycoprotein cluster of differentiation 147 (CD147) [197], thyroid hormones and TSH [84,85,86], protein kinase (PK)D3 signaling [198], and potentially the Eph ligand ephrin A3 (EFNA3) [45]. For example, hypoxia activates the PI3K-Akt-mTOR signaling cascade and thus SREBP-1 in experimental murine HCC via the Hippo pathway, which in turn is stimulated via the PGE2 receptor 4 (EP4) by PGE2 that is released from the mesenchymal tumor environment [57]. In addition, mTORC1 forms a hub for nutrient/amino acid sensing [111], which regulates SREBP-1 expression and maturation in HCC independently of PI3K and Akt [199]. The mechanisms that translate PI3K-Akt-mTOR activation into increased SREBP-1c activity are diverse and tightly cross-regulated [80, 200, 201]. This combined signaling network drives anabolic cell metabolism with effects on cell growth, proliferation, and survival and supports tumorigenesis, in addition to orchestrating a variety of other physiological and pathophysiological cellular functions [7, 8, 11, 202]. For example, mTORC1 activation leads to the phosphorylation and subsequent inactivation of cAMP response element binding protein (CREB)–regulated transcription co-activator 2 (CRTC2) [203], thereby enabling the transport of the SREBP/SCAP complex from the ER to the Golgi in the liver, dependent on component of the coat protein complex II (COPII) [204]. Moreover, mTORC1 adjusts the nuclear availability of SREBP-1 through the phosphatidate phosphatase lipin-1 [200], co-activates SREBP-1-regulatory kinases that influence maturation and stability [80], and acts on additional factors, such as the phosphorylated signal transducers and activators of transcription 5 (STAT5) [8] and the nuclear import-regulating oxidoreductase tat-interacting protein 30 (TIP30) [56].
The lipogenic function of mTOR is associated with HCC progression. Mice with liver-specific mTORC1 activation develop spontaneous HCC, which is accompanied by increased expression of SREBP-1c and lipogenic enzymes (i.e., ACC1, FASN) as well as lipid accumulation [8]. It should be noted that the predominant mechanisms by which mTOR triggers SREBP-1c signaling are cell-type- and context-dependent and that PI3K-Akt stimulates SREBP-1-mediated lipogenesis also independently of mTOR [205]. Consistent with this finding, liver-specific deletion of tuberous sclerosis 1 (TSC1), a cochaperone that negatively regulates mTORC1 signaling [206], activates mTORC1 but shows defects in SREBP-1 maturation and fails to protect against age- and diet-dependent hepatosteatosis in mice [199, 207]. Below, we describe major signaling cascades/factors (protein kinases, lipin-1, CRTC2) that are used by HCC to translate an activation of the PI3K-Akt-mTORC1-axis into SREBP-1 signaling. In addition, we highlight selected signaling cascades (CD147, thyroid hormones, EFNA3, and TIP30) that activate the PI3K-Akt pathway, induce SREBP-1 expression or activation, and have (pre)clinical potential in the context of HCC.
6.1 Protein kinases
6.1.1 Phosphoenolpyruvate carboxykinase 1 (PCK1)
PCK1 is one of the rate-limiting enzymes in gluconeogenesis and is involved in the regulation of SREBP-1 [208]. Akt phosphorylates PCK1 at serine-90 (S90) in human Huh-7 hepatoma cells, resulting in the translocation of the kinase to the ER, where PCK1 uses guanosine-5′-triphosphate (GTP) to phosphorylate Insig1 at S207 and Insig2 at S151 [209]. Phosphorylation of Insig1/2 lowers their binding affinity to sterols, weakens their interaction with SCAP, and allows the transfer of SREBP-1/2 from the ER to the Golgi, where proteolytic processing to the mature transcription factor takes place. The proliferation of HCC cells is in consequence induced, as is tumor growth in HCC-grafted mice [209].
6.1.2 Glycogen synthase kinase 3 (GSK3)
SREBPs contain a consensus phosphopeptide motif (CPD) that interacts with F-box and WD repeat domain-containing 7 (FBW7), the substrate recognition component of a complex of Skp1, Cul1, and F-box protein (SCF)-type ubiquitin ligase [210]. GSK3 phosphorylates the CPD, thereby recruiting FBW7 to SREBPs and stimulating ubiquitination and proteasomal degradation, as shown for diverse cancer cell lines including HCC cells [112, 211,212,213]. Specifically, GSK3 phosphorylates human SREBP-1 at threonine-426 (T426) and S430 [112], human SREBP-2 at S432 and S436 [112], and rat SREBP-1 at S73 [214]. Inhibition of GSK3 by Akt or the Akt-activating kinase mTORC2, therefore, stabilizes mature SREBP, enhances lipogenesis, and is associated with HCC cell proliferation and survival [6, 7, 112, 211]. In this sense, mTORC2 activates Akt by phosphorylation at S473, increases mSREBP levels, induces lipid accumulation, and promotes HCC in mice deficient in hepatic Tsc1 and phosphatase and tensin homolog (Pten), and its activity correlates with lipogenesis and HCC in patients [41]. Note that the role of GSK3 in HCC remains controversial, with several studies also suggesting GSK3 as an anti-tumoral target [215].
6.1.3 p70 S6K
Activation of PI3K-mTORC1 by insulin or growth factors stimulates SREBP-1c processing via p70 S6K [80]. p70 S6K then phosphorylates ribosomal protein S6 (RPS6) [216], which fosters HCC progression and increases the expression of lipogenic enzymes (FASN, ACLY, SCD1, 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), mevalonate kinase (MVK)) and promotes lipogenesis in human HCC cell lines, human HCC specimen, and liver of transgenic mice expressing constitutively active Akt [6]. The authors propose that the activation of the RPS6 pathway disrupts the FBW7-SREBP-1 and FBW7-SREBP-2 degradation complexes [6], which have previously been shown to initiate the ubiquitination and proteasomal degradation of nuclear SREBP-1a, SREBP-1c, and SREBP-2 [112].
6.2 Lipin-1
The phosphatidic acid phosphatase lipin-1 critically regulates lipid homeostasis, either through its enzymatic activity or by co-regulating gene transcription [217]. Active lipin-1 in the nucleus evokes nuclear eccentricity, reduces SREBP promoter activity, and decreases mSREBP abundance [200]. Phosphorylation of lipin-1 by mTORC1 prevents the nuclear entry of the phosphatase and thereby mediates the effect of mTORC1 on SREBP gene expression through undefined mechanisms, as shown for murine fibroblasts, AML12 hepatocytes, and HepG2 cells in vitro and in the liver of mice fed a Western diet [200]. Lipin-1 phosphorylation by mTORC1 and casein kinase I further allows recognition by the SCFβ-TRCP E3 ubiquitin ligase complex and subsequent degradation [218]. Interestingly, the lipin-1 promoter contains an SRE site, which explains why SREBP-1 activation upon sterol depletion enhances lipin-1 transcription in HCC cell lines [219]. Induction of lipin-1 expression by SREBP is synergistically enhanced by the transcription factor nuclear factor Y (NF-Y) [219].
6.3 CRTC2
CRTC2 belongs to a family of CREB co-activators and regulates glucose and lipid metabolism in the liver and other tissues [128, 204]. As shown in the liver of mice at chow and high-fat diet, CRTC2 is phosphorylated at S136 by mTORC1 and mediates the effect of mTORC1 on SREBP-1 processing [128, 204]. Specifically, CRTC2 competes with SEC23 homolog A (Sec23A) to interact with Sec31A within the COPII complex, which is required for the transport of SREBP-1 from the ER to the Golgi [204]. Hepatic overexpression of the mTOR-defective mutant CRTC2(S136A) counteracts the unleashed SREBP-1 signaling in mouse liver on a high-fat diet and improves insulin sensitivity [204]. These findings indicate that CRTC2 plays a central role in controlling SREBP-1 signaling and lipid homeostasis in the liver. Less well understood is the extent to which CRTC2 regulates SREBP-1 and contributes to metabolic reprogramming in HCC. A recent report indicates that CRTC2 expression is induced in mouse liver by high-fat diet and activates the mTOR pathway by increasing miR-34a levels, reducing SIRT1-dependent deacetylation and downregulating TSC2 [220].
6.4 TIP30 (HTATIP2, CC3)
The tumor suppressor TIP30 belongs to the short-chain dehydrogenases/reductases (SDR) family of more than 2000 NAD(H)/NADP(H)-dependent oxidoreductases and is involved in the regulation of cancer cell metabolism, survival, growth, and metastasis [221]. In the context of HCC, TIP30 deficiency has been shown to promote lipogenesis and cell proliferation in HCC cell lines and in a mouse xenograft model by enriching SREBP-1 in the nucleus through the Akt-mTOR pathway and inducing SREBP-1 and subsequent target gene expression [56]. Accordingly, TIP30-knockout mice are more prone to spontaneously develop HCC and other tumors as compared to wild-type mice [222]. The critical role of SREBP-1 regulation by TIP30 in tumorigenesis is further supported by the finding that TIP30 and SREBP-1 levels are inversely correlated in tumor samples from HCC patients [56]. Such an association is not observed for the surrounding non-tumorous area.
6.5 Selected PI3K/Akt-regulatory pathways
Hepatic PI3K-Akt activation is not limited to insulin and growth factors [223]. Below, we exemplarily highlight three pathways that trigger the PI3K-Akt cascade and have been shown to regulate SREBP-1 in HCC.
6.5.1 CD147
The transmembrane glycoprotein CD147 is overexpressed in many human cancers, including HCC, and has attracted interest as a pharmacological target for the treatment of HCC [224]. Deletion of CD147 reduces the aggressiveness (i.e., tumor growth rate and metastasis) of human SMMC-7721 hepatocarcinoma cells in mouse xenografts, which is compensated by SREBP-1 overexpression [197]. Specifically, CD147 induces the expression of lipogenic enzymes (ACC1, FASN) by activating the AKT-mTOR-SREBP-1 pathway and represses enzymes involved in fatty acid β-oxidation (carnitine palmitoyltransferase 1 (CPT1A), peroxisome acyl-coenzyme oxidase 1 (ACOX1)) by activating p38 and downregulating PPARα [197]. In addition, N-acetylglucosaminyltransferase V (GnT-V)–mediated N-glycosylation of CD147 enhances its interaction with integrin β1 and stimulates HCC metastasis [225].
6.5.2 Thyroid hormones
The thyroid hormone 3,5,3′-triiodo-L-thyronine (T3) increases the expression of SREBP-1, apparently by binding to the integrin receptor αvβ3 and activating the PI3K-Akt-mTORC1 pathway in HepG2 cells, as suggested by inhibitor studies [86]. Protein kinase C-alpha (PKC-α) seems to act as a reverse regulator, dampening the T3-mediated upregulation of SREBP-1 [86]. 3,5-Diiodo-L-thyronine (T2), a precursor and by-product of thyroid hormone biosynthesis, instead activates p38 and extracellular signal-regulated kinase 1/2 (ERK1/2) and suppresses Akt signaling, which inhibits SREBP-1 maturation and promotes apoptosis in HepG2 cells [85].
6.5.3 Ephrin A3 (EFNA3)
Ephrins are membrane-bound ligands that bind to Eph receptors, which form the largest family of RTKs [226]. Signaling by Eph-ephrin can be forward, reverse, or bi-directional and does not necessarily require receptor-ligand interaction [226]. The Eph-ephrin system is involved in various developmental processes [226, 227]; is implicated in HCC metabolic plasticity, HCC malignancy, tumor progression, and aggressiveness (with mixed results regarding the directionality of the regulation) [45, 228, 229]; and inhibits PI3K-Akt-signaling [228]. EFNA3 is abundantly expressed in clinical HCC samples, and EFNA3 levels correlate with tumor aggressiveness [45]. EFNA3 expression is induced in HCC cell lines by HIF-1α under hypoxic conditions [45], which mimics the tumor microenvironment in hyperproliferative HCC in vivo [230, 231]. Through forward activation of EPH receptor A2 (EphA2), EFNA3 induces SREBP-1 maturation and expression of the SREBP-1 target gene ACLY, which mediates the self-renewal (cancer stemness–promoting) activity of the ephrin A3-EphA2-axis through metabolic rewiring [45]. Whether SREBP-1 activation is dependent on PI3K-Akt-signaling has not been explicitly investigated.
6.6 Pharmacological interference
A large variety of small molecules interfere with SREBP-1 signaling by targeting SREBP-1 expression, maturation, degradation, post-translational modification, and upstream signaling cascades, such as PI3K, Akt, or mTOR. However, only a limited number of drugs, drug candidates, and natural products have been explicitly reported to inhibit SREBP-1 activity in HCC and HCC-related liver pathologies (Table 1). For example, bergapten (25–50 mg/kg/day, intraperitoneal administration (i.p.)), a furanocoumarin widely found in plants, suppresses the growth of N-nitrosodiethylamine (NDEA)-induced HCC in rats [58]. The anti-tumoral effects were correlated with increased LXRα/β expression (see the “LXR” section), suppressed fatty acid biosynthesis, impaired activation of the PI3K-Akt-SREBP-1 pathway, and reduced cholesterol uptake, likely due to a decrease in low-density lipoprotein receptor (LDLR) and an increase in cholesterol efflux through the upregulated ATP-binding cassette subfamily A member 1 (ABCA1) [58]. Another example is the anti-obesity drug orlistat, which increases the expression of PTEN, thereby inactivating Akt signaling and inhibiting SREBP-1 activation in vitro and in vivo. As a result, lipogenesis (as indicated by FASN expression) and cell proliferation are reduced in human hepatoma cell lines (20–50 μM orlistat), and steatosis and HCC progression are inhibited in rodent models (150–300 mg orlistat/kg/day; i.p.; 10–45 mg orlistat/kg/day, p.o.) [40, 124,125,126].
7 AMPK
The serine-threonine kinase AMPK is an energy-sensor kinase that inhibits energy-consuming metabolic processes, including lipogenesis, and is downregulated in HCC patients [6, 107]. AMPK negatively regulates SREBP-1 transcription, proteolytic processing, nuclear translocation, and activation through multiple mechanisms, both directly through phosphorylation and indirectly by repressing pathways involved in SREBP-1 activation such as mTOR signaling [107]. Accordingly, hepatocyte-specific deletion of AMPK induced a mild steatotic phenotype in mice fed a high-fat/high-sucrose diet as compared to wild-type littermates [135]. In the context of liver physiology and HCC, AMPK activators such as metformin and 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) (Table 1) have been shown to negatively regulate hepatic transcription and processing of SREBP-1 and contribute to reduced HCC cell proliferation and liver tumorigenesis in mice [47, 79, 129,130,131].
Here, we briefly summarize the major mechanisms by which AMPK is considered to counteract SREBP-1 signaling in the liver. Firstly, AMPK phosphorylates S372 of SREBP-1c in mouse liver and inhibits SREBP-1c cleavage and nuclear translocation [134]. Insig1 is also directly phosphorylated by AMPK at T222, which ablates its interaction with gp78, stabilizes Insig1, and inhibits SREBP-1 cleavage and processing in mouse liver [135]. AMPK negatively regulates mTORC1 [107], which induces SREBP-1 signaling [201], and this mechanistic link has been proposed to contribute to the anti-tumoral activity of sorafenib in hepatocarcinoma cells [143]. By suppressing the biogenesis of circPRKAA1, a circRNA transcribed from the AMPK α1 subunit that interacts with Ku proteins (Ku80 and Ku70), AMPK decreases the stability of mSREBP-1 and reduces the development of tumors, including HCC [232]. Other proposed mechanisms by which AMPK interferes with SREBP-1 signaling include decreased cholesterol/LXR ligand biosynthesis, the inhibition of LXR-dependent SREBP-1c promoter activity, and diminished cleavage (maturation) of pSREBP-1c, which is likely to contribute to the suppressive effect on SREBP-1c transcription [233, 234]. A role for AMPK in suppressing SREBP-1 activity has also been described for the lipogenic activity of human HCC-associated protein TD26 (TD26). This oncoprotein is highly expressed in HCC tumor tissue (as compared to matched normal tissue), and its levels correlate with tumor size and poor prognosis in HCC patients [235]. TD26 interacts with SREBP-1 through its C-terminal domain and lowers AMPK-mediated suppression of SREBP-1 activity, thereby elevating lipogenesis and enhancing HCC cell proliferation and tumor growth [235]. In addition, AMPK enhances the activity of DNA methyltransferase 3A (DNMT3A), which maintains 5-methylcytosine (5mC) modifications at the thymine DNA glycosylase (TDG) promoter and diminishes TDG expression [79]. TDG is transactivated by c-Myc [79] and works in concert with members of the ten-eleven translocation (TET) family to oxidize 5-methylcytosine and cleave the product 5-carboxylcytosine (5caC) [236] from the SREBP-1 promoter (base-excision repair), among others, in hepatocarcinoma cells [79].
7.1 Regulation of AMPK and link to ferroptosis
AMPK is regulated by a variety of signaling pathways [237]. Here, we briefly outline selected strategies used by HCC to control SREBP-1 signaling via AMPK and highlight the emerging link to ferroptosis, an alternative cell death program to apoptosis that is driven by excessive membrane peroxidation [113, 238].
The AMPK-SREBP-1c axis plays a central role in linking glucose and lipid metabolism in HCC [33]. For example, elevated exocellular lactate levels and lactate import activate the SREBP-1-SCD1 axis by repressing AMPK and promoting resistance to ferroptosis inducers [37]. Lactate accumulation is a hallmark of cancer cells, including HCC, and is attributed to the Warburg effect, which describes the preference of cancer cells for aerobic glycolysis over oxidative phosphorylation for energy production [239]. AMPK, SREBP-1c, and its target genes FASN and SCD1 determine susceptibility to membrane peroxidation and have been recognized as key players in ferroptosis [74, 240,241,242,243,244,245,246]. Accordingly, lactate depletion or inhibition of lactate import via the hydroxycarboxylic acid receptor 1 (HCAR1)/monocarboxylate transporter 1 (MCT1) impairs ATP generation and activates AMPK to repress SCD1, but also upregulates long-chain fatty acid CoA ligase 4 (ACSL4) [37], a lysophospholipid acyltransferase that preferentially channels PUFAs into membrane phospholipids [247]. The authors propose that both effects contribute to the induction of ferroptosis in HCC cells. Supplementation with SCD1 products, i.e., the MUFAs oleic acid or palmitoleic acid, actually compensates for the cytotoxicity induced by HCAR1/MCT1 suppression [37]. Note that lactate increases the levels of HCAR1, which is upregulated in tumors of HCC patients compared to adjacent tissues [37]. Together, lipid metabolic adaptations through the AMPK-SREBP-1c system essentially regulate ferroptosis sensitivity in HCC, as recently suggested for primary mouse embryonic fibroblasts and several cancer cell lines [37, 240, 248].
Another mechanism by which AMPK enhances ferroptosis sensitivity of HCC is related to branched-chain amino acid aminotransferase 2 (BCAT2), which was identified as a ferroptosis suppressor in a kinome CRISPR/Cas9-based screen in HepG2 cells [98]. BCAT2 catalyzes the reversible transamination of branched-chain amino acids and α-ketoglutarate, converting them to the corresponding α-keto acids and glutamate [98]. Note that the availability of glutamate is essential to keep ferroptosis at bay [238]. Accordingly, HCC cells are protected from lipid peroxidation when cellular glutamate levels and glutamate release are maintained [98]. Feedback loops seem to exist: the induction of ferroptosis in HepG2 cells has been proposed to trigger ferritinophagy and increase ROS levels, thereby activating AMPK, inhibiting SREBP-1, and downregulating BCAT2 [98].
There are also emerging links between AMPK and the ubiquitin-conjugating system, which interestingly has recently been implicated in the regulation of ferroptosis [249, 250]. In HCC cells, ubiquitin-conjugating enzyme E2 O (UBE2O) has been shown to confer malignant features, such as cell growth, migration, and invasion, by decreasing AMPKα2 stability and promoting the mTORC1 pathway [249]. UBE2O is overexpressed in HCC compared to adjacent normal tissues, and high UBE2O levels correlate with worse clinical outcomes in HCC patients [249]. Future studies are required to elucidate whether the UBE2O-dependent degradation of AMPKα2 along with mTORC1 activation effectively activates SREBP-1 in HCC and whether such regulation is functional in terms of ferroptosis sensitization and tumorigenesis.
Given the important role of inflammation and immune surveillance in HCC initiation and progression [230, 251,252,253], it is also noteworthy that HCC upregulates the expression of pro-inflammatory cytokines [254, 255]. TNFα and related cytokines promote tumor growth under certain conditions by creating a pro-inflammatory tumor microenvironment and/or initiating pro-survival signaling cascades [256, 257] and may also promote lipogenesis by interfering with AMPK activation (and other pathways) and relieving the repression of the mTORC1-SREBP-1c pathway, as shown for HCC cells [258, 259]. SREBP-1 itself has a dual role in inflammation in the tumor microenvironment, acting either to promote inflammation [38] or to resolve inflammation [39].
7.2 Pharmacological interference
AMPK activators such as metformin [47, 79, 129,130,131,132,133,134,135,136,137,138,139,140,141, 260,261,262,263,264,265,266,267,268] and AICAR [98, 132, 133, 135, 139, 142, 262, 269,270,271] have been extensively studied in liver pathologies, including HCC, and have been shown to suppress lipogenesis and/or tumorigenesis in pre-clinical and clinical studies (Table 1). Note that metformin activates AMPK in the liver through direct interaction with PEN2, a subunit of γ-secretase, resulting in inhibition of v-ATPase and activation of AMPK [264], but also has AMPK-independent activities [266, 272, 273]. Although a negative regulation of SREBPs is widely expected, only a fraction of these studies provided experimental evidence to support that AMPK inhibitors interfere with SREBP-1 signaling in HCC [47, 129, 131, 135, 136].
The multikinase inhibitor sorafenib is in clinical use for the treatment of HCC [274, 275]. Among many other activities, sorafenib has been shown to activate the cellular energy sensor AMPK by decreasing ATP production [143]. Subsequent phosphorylation of TSC2 at S1387 [276] and the mTORC1 subunit Raptor at S722 and S792 inhibits mTORC1-mediated SREBP-1 activation [277], suppresses the expression of the SREBP-1 target gene SCD1, and reduces the availability of MUFAs in Huh7 cells [143, 278]. Interestingly, sorafenib-resistant HCC cell lines highly express programmed death ligand 1 (PD-L1), which activates the PI3K-Akt-SREBP-1 axis in HCC cell lines and installs features that are characteristic of EMT [72]. Accordingly, high levels of SREBP-1 in tumor tissue of HCC patients receiving sorafenib correlate with poor prognosis [66], which suggests that SREBP-1 contributes to the resistance of HCC cells to sorafenib. In support of this hypothesis, the knockdown of SREBP-1 synergizes with the tumor-suppressive activity of sorafenib against HCC [66].
Diverse natural products have been reported to activate AMPK and suppress SREBP-1 in HCC (Table 1). For example, the polyphenol resveratrol (30 mg/kg/day, p.o.) protects against hepatitis B virus (HBV)–induced fatty liver and HCC (incidence reduced from 80 to 15%) in HBV X protein (HBx) transgenic mice. Resveratrol represses major enzymes in de novo fatty acid biosynthesis, i.e., ACC and FASN, seemingly by inhibiting LXRα and downregulating SREBP-1 and PPARγ and activating AMPK and SIRT1 [147]. In addition, resveratrol promotes transient liver regeneration and stimulates the cellular antioxidant response, as indicated by elevated intracellular glutathione levels [147]. Along these lines, piceatannol (3,3′,4,5′-tetrahydroxy-trans-stilbene) (at 100 μM), a stilbenoid metabolite of resveratrol, suppresses the mRNA expression of SREBP-1c and PPARγ, while inducing PPARα in HepG2 cells, which is associated with an apparent shift from anabolic to catabolic fatty acid metabolism [191]. Based on previous studies on resveratrol and piceatannol suggesting activation of AMPK in liver disease [279], the authors propose that the two polyphenols employ similar AMPK-dependent mechanisms to attenuate lipid accumulation [191]. Other examples include isoquercitrin (at 10–200 μM) [148,149,150], the tetracyclic triterpenoid limonin (50–200 μM; 50 mg/kg/day, p.o.) [155], and curcumin (1–10 μM; 80–100 mg/kg/day, p.o.) [151, 152], which promote the phosphorylation of AMPKα, decrease SREBP-1 expression, downregulate lipogenic proteins or interfere with their activation, and reduce total lipid content in HCC cell lines [151]. Inhibitor studies for isoquercitrin confirmed a functional link between AMPK and ACC and adiponectin receptor 1 (AdipoR1) expression [148]. Furthermore, the pentacyclic triterpenoids tormentic acid (0.06–0.12 g/kg/day, p.o.) and betulinic acid (5–10 mg/kg, p.o.) and the pentacyclic triterpene saponin pedunculoside (5–30 mg/kg/day, p.o.) inhibit high-fat diet–induced hepatic lipid accumulation [127, 160, 169] and hyperlipidemia [127, 169] in rodents along with increasing AMPK phosphorylation and reducing SREBP-1 expression. Similar mechanisms seem also to apply to distinct plant extracts with hepatoprotective properties. For example, mulberry anthocyanin extract (at 300–500 μg/ml) and fermented Rhus verniciflua Stokes extract (at 200–400 μg/ml) inhibit triglyceride and/or cholesterol accumulation in HepG2 cells that were overloaded with oleic acid [280, 281], which has been ascribed to enhanced AMPK phosphorylation and decreased SREBP-1 expression.
8 PKA
The serine-threonine kinase cAMP/cAMP-dependent protein kinase A (PKA) is regulated by various stimuli that activate G-protein-coupled receptors (GCPRs) or interfere with the adenylate cyclase/phosphodiesterase system to modulate the concentrations of the second messenger cAMP, which activates PKA [83]. Among many other functions, PKA is a critical signaling kinase in the control of glucose and lipid metabolism, converging signal transduction by GCPR ligands, including insulin and glucagon [83]. PKA plays a central role in suppressing lipogenesis, among others, through pathways that negatively regulate SREBP-1 [83]. By directly phosphorylating human SREBP-1a at S338 and SREBP-1c at S314, PKA suppresses transcriptional activity [282]. In addition, PKA represses the expression of SREBP-1c by activating the FXR or SIRT1 [83] and by phosphorylating AMPKα1 at S173, which inactivates the kinase by impeding T172 phosphorylation [283]. Of relevance to liver metabolism and HCC, glucagon triggers cAMP generation and has opposite effects compared to insulin, among others, by preventing insulin-induced SREPB-1c expression and maturation [80]. cAMP-activated PKA phosphorylates SREBP-1a, thereby attenuating their DNA occupancy in cell-free assays and in HepG2 cells [284]. On the other hand, PKA phosphorylates PPARα, which induces SREBP-1c transcription and likely explains why hepatic lipid accumulation is increased in TSH-deficient mice on chow and high-fat diet [84]. Conclusively, cAMP-PKA exerts both lipogenesis-promoting and lipogenesis-inhibiting activities by regulating SREBP-1 in HCC depending on the stimulus and context.
9 LXR
LXRs (LXRα and LXRβ) are nuclear hormone receptors that form heterodimers with retinoid X receptor (RXR) and are activated by oxysterols and several exogenous ligands [285]. By transactivating metabolic drivers, such as SREBP-1c and ABCA1, LXRs regulate cholesterol homeostasis and lipogenesis [285, 286]. While LXRβ is ubiquitously expressed, LXRα is most abundant in metabolic tissues, including liver [285]. Various metabolic signaling pathways that regulate SREBP-1 signaling cross-talk with LXRs. For example, PPARα and PPARγ (see the “PPARs” section) directly bind to and transactivate the LXRα promoter [287, 288], and PI3K-Akt-mTORC1 activation by insulin or growth factors (see the “PIAK-Akt-mTOR axis” section) stimulates SREBP-1c transcription through diverse mechanisms, some of which involve LXR [77, 289]. Endogenous ligands that antagonize LXR activity include PUFAs, such as arachidonic acid and docosahexaenoic acid [290] (see the “PUFAs” section). Note that LXRs are upregulated in various cancers and that LXR agonists have been shown to counteract tumorigenesis and metastasis in experimental cancer models [285].
In the context of HCC, LXRα/β and its heterodimeric partner RXR target two LXR binding elements (LXREs) at the SREBP-1c promoter and conjointly activate transcription, as shown by LXR and RXR transfection and pharmacological approaches using the natural LXR ligand 22(R)-hydroxycholesterol and the RXR ligand 9-cis-retinoic acid [10, 291]. Accordingly, both total and nuclear SREBP-1c levels increase along with FASN expression. The observation that LXR activation induces SREBP-1 mRNA expression and increases mSREBP-1 levels was independently confirmed in HepG2 cells using the selective LXR ligand T0901317 [292].
The activity of LXR is inhibited by a short heterodimer partner (SHP) in rat hepatoma McA-RH7777 cells, likely through suppressive heterodimerization that reduces SREBP-1 expression [97]. SHP is an atypical member of the nuclear hormone receptor family, which possesses a putative ligand-binding domain but lacks a conventional DNA-binding domain [293]. Activation of FXR induces the expression of SHP, which explains the triglyceride-reducing activity of natural FXR ligands such as bile acids [97]. In fact, a bile acid (cholic acid)–rich diet decreases SREBP-1 promoter activity, diminishes hepatic mRNA expression of SREBP-1c and other lipogenic genes, and lowers plasma and hepatic triglyceride levels in mouse models of hypertriglyceridemia [97].
10 PPARs
The three PPAR isotypes PPARα, PPARβ/δ, and PPARγ form heterodimeric complexes with RXR and act as nuclear receptors for endogenous ligands, such as fatty acids and their derivatives [294, 295]. PPARα is highly expressed in the liver and is targeted by fibrates, lipid-lowering drugs used to treat dyslipidemia, whereas PPARγ is the target of thiazolidinediones, anti-diabetic drugs that sensitize extrahepatic tissues to insulin [295]. PPARs have both positive and negative effects on the SREBP-1 pathway [296, 297]. For example, PPARα induces SREBP-1c transcription by binding directly to the DR1 element at the SREBP-1c promoter in human liver [288], but also upregulates Insig2α expression during fasting, thereby suppressing SREBP-1c processing [297]. PPARδ promotes Insig1 expression by binding upstream of the transcription initiation site and suppresses SREBP-1 proteolytic processing in the liver of obese diabetic mice [296]. With respect to HCC, both PPARα and PPARγ activation have been shown to reduce nuclear SREBP-1 availability, inhibit SREBP-1 target gene expression, and decrease triglyceride production in rat hepatoma Fao cells [298]. In the previous section “PKA,” we have already mentioned the regulation of SERBP-1 by PPARs as part of or in parallel with other SREBP-1-regulatory pathways (cAMP-PKA, LXRα).
11 PGC-1
The induction of SREBP-1c (and to a lesser extent SREBP-1a) transcription by LXR is amplified (in HepG2 cells) by peroxisome proliferator-activated receptor γ co-activator 1α (PGC-1α), which acts as a co-activator of LXR [299]. PGC-1β also co-activates SREBPs, contributing significantly to the SREBP-dependent lipogenic gene expression in mouse H2.35 hepatoma cells and rat liver [300]. PGC-1β physically interacts with SREBP-1c through a specific domain (amino acids 350–530) that is absent in PGC-1α. Despite the induction of lipogenic SREBP-1c target genes in PGC-1β-transgenic rats, hepatic triglyceride and cholesterol levels were markedly decreased, seemingly due to the co-activation of LXR by PGC-1β and stimulation of lipoprotein secretion [300].
Through a PGC-1-dependent mechanism, pharmacological inhibition of p38 increases insulin-induced expression of SREBP-1 target genes in primary hepatocytes [82]. p38 inhibition also induces triglyceride accumulation in mouse liver on high-fat diet and reverses the decrease in triglyceride levels along with the expression of SREBP-1 and lipogenic target enzymes, as shown in glucagon-treated primary hepatocytes (see the “PKA” section) and in fasting mouse liver [82]. Mechanistically, p38 attenuates the insulin-induced increase as well as the fasting- and glucagon-induced decrease in PGC-1β transcription [82], which inhibits the co-activation of SREBP-1 transcription [300]. Together, p38 negatively regulates SREBP-1-dependent hepatic lipogenesis through the PGC-1β-SREBP-1c pathway. The regulatory function of p38 on SREBP-1 promoter activity was confirmed in mouse Hepa1c1c7 hepatoma cells [82].
12 STAT
Janus kinase (JAK)/STAT inhibitors have been extensively studied in the context of liver pathology and are currently under clinical investigation for the treatment of HCC [301]. JAKs are associated with various cytokine and growth factor receptors and, when activated, phosphorylate intracellular receptor domains to recruit STAT proteins (STAT1-6). STATs are phosphorylated by JAKs, dimerize, and act as transcription factors to regulate the expression of specific genes involved in immunoregulation, inflammation, cell growth, survival, differentiation, and adipogenesis [301]. They are expressed in a cell type–dependent manner and induce both overlapping and unique responses. Cross-talk exists between JAK-STAT signaling and various pathways at the intersection of metabolic control and cancer, including the PI3K-Akt-mTOR axis, transforming growth factor β (TGFβ) signaling, and Notch signaling [301].
The functional link between STAT signaling and SREBP-1 is still emerging. Of relevance to HCC, mTORC1 interacts with and phosphorylates STAT5, which then binds to the SREBP-1 promoter and stimulates SREBP-1 transcription [8]. Accordingly, liver-specific activation of mTORC1 increases lipogenic gene expression and spontaneous tumorigenesis in mouse liver [8], and high levels of SREBP-1 and phospho-STAT5 are associated with poor survival in HCC patients [8]. Furthermore, liver-specific STAT5 knockout in mice induces steatosis by upregulating SREBP-1 and PPARγ signaling and simultaneously activating the c-Jun N-terminal kinase 1 (JNK1)-STAT3 pathway [87]. When liver-specific STAT5 deficiency is combined with glucocorticoid receptor deletion, fatty liver develops into HCC [87]. This malignant transformation is ascribed to lipid mobilization from adipose tissue and aggravated hepatic lipid accumulation along with insulin resistance and increased expression of the pro-inflammatory cytokine TNF-α [87]. Another study found that TNF-α and insulin upregulate proprotein convertase subtilisin/kexin type 9 (PCSK9) in HepG2 cells dependent on suppressor of cytokine signaling 3 (SOCS3)-JAK-STAT3 [92]. Overexpression of SOCS3 induces Akt phosphorylation and increases the expression of SREBP-1c, lipogenic genes, and ApoB, without affecting cholesterol biosynthesis [92]. Whether PCSK9 contributes to the regulation of SREBP-1 in addition to its role in insulin resistance has not been investigated. ROS-scavenging and inhibitor studies in high glucose–challenged HepG2 cells suggest that PCSK9 expression is induced by ROS-dependent enrichment of nuclear SREBP-1 (but not SREBP-2) [302]. Indeed, oxidative stress activates SREBP-1c and induces lipid accumulation in HepG2 cells [303], and an SRE binding site exists on the PCSK9 promoter [81]. The authors further propose that PCSK9 elevates extracellular levels of LDL cholesterol (LDLc), which protects against sorafenib-induced HCC cell death [302].
Notably, the hepatoprotective flavonolignan silibinin from Silybum marianum inhibits STAT3 activation and suppresses SREBP-1-mediated lipid accumulation in endometrial carcinoma cells and tumors [304, 305]. Although a functional link between STAT3 and SREBP-1 signaling has not been confirmed for the anti-tumoral effect of silibinin, we consider such a mechanism likely to contribute to the proposed anti-lipogenic and HCC-preventive activities of the natural extracts.
13 MYC
The proto-oncogenic transcription factor Myc is induced in response to mitogenic stimuli; regulates genes involved in cell metabolism, growth, and proliferation [306, 307], among them SREBF1; and is frequently overexpressed in cancer, including HCC [306, 307]. Direct binding of Myc to two recognition sites on the SREBP-1 promoter has been confirmed by chromatin immunoprecipitation [308]. Furthermore, N-Myc, a member of the Myc protooncogene family, induces SREBP-1 expression (along with the reprogramming of other metabolic pathways) dependent on the Myc superfamily transcription factor MondoA, which acts as a nutrient sensor [309]. Myc and SREBP-1 cooperate in the expression of lipogenic target genes and drive lipogenesis and tumorigenesis in several cancer models [310]. While Myc and SREBP-1 have been independently described to promote HCC in different systems [7, 10, 11, 306, 307], studies demonstrating that Myc induces SREBP-1 expression in HCC are rare [310]. Insulin-induced activation of c-Myc has been reported to transactivate TDG [79], which mediates hypomethylation of the SREBP-1 promoter, promotes lipogenesis, and induces the proliferation of hepatocarcinoma and other cancer cell lines [79]. In further support of Myc regulating SREBP-1 in HCC, the acyl-CoA synthetase isoenzyme ACSL4 was recently shown to promote SREBP-1 expression by stabilizing Myc in hepatoma cells [308, 311], as described in the “PUFAs” section.
14 Kinases that directly phosphorylate SREBP-1
Diverse kinases phosphorylate and thereby regulate the activity, subcellular localization, maturation, and stability of SREBP-1 [134, 214, 284]. In the context of HCC, SREBP-1 has been reported to be directly phosphorylated by GSK3, AMPK, and PKA [134, 214, 284], as discussed in the “Protein kinases” section, “AMPK” section, and “PKA” section. mSREBP-1 is also directly phosphorylated at S439 and thereby stabilized by the cyclin-dependent kinase 1 (Cdk1)/cyclin B complex [312], which coordinates G2/M progression [313]. The authors conclude that this phosphorylation might help preserve basal lipogenesis during and beyond mitosis to support cell growth and proliferation [312]. Another kinase linking glucose metabolism to SREBP-1 activation is the pyruvate kinase isoenzyme M2 (PKM2), which catalyzes the final step of glycolysis and is overexpressed in different cancer types, including HCC, but also has complex functions related to redox homeostasis and inflammation [314,315,316,317,318,319]. PKM2 phosphorylates mSREBP-1a at T59, thereby promoting interaction with PKM2, stabilizing mSREBP-1a, stimulating HCC cell proliferation, and correlating with poor prognosis of HCC patients [181]. Notably, Zhao et al. were able to block the interaction of PKM2 and mSREBP-1a and thereby attenuate lipogenic gene expression using a competitive peptide inhibitor corresponding to the amino acid sequence of SREBP-1a from 43-56 [181].
15 Histone acetyltransferases and sirtuins
The expression of SREBP-1 is controlled by post-translational acetylation, with the histone acetyltransferase CREB-binding protein/E1A binding protein p300 (CBP/p300) and sirtuins (SIRT), a family of NAD+-dependent deacetylases, dominating the regulation of SREBP-1 in HCC [182, 320, 321]. Histone acetyltransferases CBP/p300 activate the transcriptional activity of SREBP-1c by acetylating the transcription factor at lysine-289 (K289) and K309, which promotes the accessibility to target gene promoters and enhances SREBP-1c stability by suppressing ubiquitination-dependent degradation [320].
The seven members of the SIRT family are class III histone deacetylases that differ in subcellular localization and functions related to cell metabolism, cancer, and many other physiological and pathophysiological processes [322]. SIRT1 directly interacts with SREBP-1c, decreases the acetylation at K289 and K309, and inhibits the lipogenic activity of SREBP-1c in mouse liver by reducing its stability and occupancy at target gene promoters [182]. The interaction of SIRT1 with SREBP-1c is enhanced during fasting and reduced during feeding (insulin/glucose), reflecting changes in SREBP-1c acetylation [182]. SIRT1 is regulated by manifold pathways and cellular processes, such as the DNA damage and oxidative stress response [322, 323]. In HCC, activation of SIRT1 is vital for cancer progression and has been closely linked to mitochondrial fission [321, 324], a tightly regulated process used to meet the metabolic demands of the cell [325]. Increased mitochondrial fission decreases cellular NAD+ levels and SIRT1 activity in HCC cells, which increases the acetylation of SREBP-1 and PGC-1α [321]. As a result, lipogenic SREBP-1c target enzymes (FASN, ACC1, and ELOVL6) are upregulated, while the expression of enzymes involved in fatty acid oxidation (CPT1A and ACOX1, which are under the control of PGC-1α/PPARα) is inhibited. This reprogramming of lipid metabolism is associated with lipid accumulation (fatty acids, triglycerides, phospholipids) and increased metastasis [321].
SIRT6 inhibits the transcription of SREBP-1 and lipogenic target genes and seems to be regulated in HCC by the nuclear envelope protein sperm-associated antigen 4 (SPAG4) [125]. Since SPAG4 binds to lamin A/C with higher affinity than SIRT6, the authors speculate that the competitive binding of SPAG4 to lamin A/C releases SIRT6 and increases the expression, nuclear translocation, and transcriptional activity of SREBP-1. SPAG4 is highly expressed in HCC, and its levels correlate with poorer survival in HCC patients [125]. In support of a functional role of SREBP-1 in SPAG4-driven HCC progression, an interference with SREBP-1 signaling by orlistat attenuates tumor growth in mouse xenografts and synergizes with sorafenib [125]. In addition, SIRT6 activation by fluvastatin (an HMG-CoA reductase inhibitor) increases SIRT6 expression and stimulates inhibitory SREBP-1 phosphorylation via AMPK in HepG2 cells, thereby decreasing lipid biosynthesis, as expected [170].
SIRT7 binds to SREBP-1 and maintains the expression of isocitrate dehydrogenase 1 (IDH1) in a SREBP-1-dependent but SIRT7 deacetylase activity-independent manner in HCC cell lines [34]. IDH1 utilizes NADP+ and converts isocitrate to α-ketoglutarate and NADPH by oxidative decarboxylation but also catalyzes the reverse reaction and contributes to lipid biosynthesis from glutamine in several cancers [326,327,328,329,330]. Silencing of SIRT7 accordingly reduces lipogenesis and gluconeogenesis, as expected from interference with the (seemingly stimulatory) interaction of SIRT7 with SREBP-1, which sustains cellular IDH1 levels [34].
16 Further post-translational modifications of SREBP-1
In addition to phosphorylation (“Kinases that directly phosphorylate SREBP-1”), acetylation (“Histone acetyltransferases and sirtuins”), and ubiquitination (“PI3K-Akt-mTOR axis”), SREBP-1 has been reported to undergo methylation and conjugation with arginine methyltransferase 5 (PRMT5) and neddylation by NEDD8-conjugating E2 ligase (UBC12) in HCC cells [65, 331]. PRMT5 symmetrically dimethylates SREBP-1a at arginine-321 (R321) in HepG2 cells, which prevents destabilizing phosphorylation at S430 by GSK3β and thereby attenuates the recruitment of the ubiquitin ligase FBW7 [331]. Thus, SREBP-1a methylation promotes lipogenesis and HCC cell growth/proliferation in vitro and in vivo and correlates with tumor progression in HCC patients [331].
NEDD8 (neural precursor cell expressed developmentally downregulated protein 8) is a ubiquitin-like protein that is covalently attached to substrate proteins by the ubiquitylation machinery and regulates various signaling pathways besides cullins and other non-cullin ubiquitin E3 ligases [332]. Neddylation of SREBP-1 is mediated by UBC12 in HepG2 and grafted SK-Hep1 cells (a human endothelial-like cell line derived from HCC) [65]. Conjugation with NEDD8 stabilizes SREBP-1 (which is less ubiquitylated) and correlates with poor metastatic tumor prognosis in HCC. Selective inhibition of neddylation by MLN4924 (an inhibitor of Nae1) (30 mg/kg, twice daily, subcutaneous administration (s.c.)) in SK-Hep1 mouse xenografts indeed reduces SREBP-1 levels, lipogenesis, and tumor growth while inducing cell death [65]. Note that proteomic and electron microscopic data suggest that SK-Hep1 cells are of endothelial and not hepatocytic origin, although they have been widely used as a cell model for HCC [333].
17 Sterols and oxysterols
As described in the “SREBP-1” section, SREBP-1/2 maturation responds to cellular metabolic demands and is induced by insufficient availability of sterols, such as cholesterol or 25-hydroxycholesterol [99, 334,335,336]. In the context of HCC, dietary cholesterol suppresses tumorigenesis by reducing SCAP-dependent de novo lipogenesis, whereas cholesterol deprivation has the opposite effect, as shown for DEN-induced HCC in mice [55]. Accordingly, overexpression of the cholesterol-biosynthetic enzyme NAD(P)-dependent steroid dehydrogenase-like (NSDHL) decreases the levels of active nuclear mSREBP-1 in HCC cell lines, whereas silencing of the enzyme increases mSREBP-1 activation [73]. The authors suggest that the SREBP-1-dependent expression of the immunosuppressive cytokine TGFβ contributes to resistance to immune checkpoint inhibitors. Indeed, immune checkpoint inhibitors activate tumor antigen–specific T cells to produce IFN-γ [337], and IFN-γ decreases NSDHL expression and stimulates SREBP-1 maturation in HCC with subsequent release of TGFβ [73]. Activation of SREBP-1/2 is also closely related to the activity of other steroid biosynthetic enzymes, including cholesterol 7α-hydroxylase (CYP7A1) [97, 335] and oxysterol 7α-hydroxylase (CYP7B1) [338], which catalyze the first step in bile acid biosynthesis by converting cholesterol to 7α-hydroxycholesterol [339] and metabolize 25- and 27-hydroxycholesterol, respectively [339]. For example, overexpression of CYP7A1 in rat hepatoma McArdle cells lowers cellular cholesterol levels, enhances SREBP-1 activation, and induces the expression of enzymes involved in fatty acid and sterol biosynthesis, along with increased cellular lipid synthesis and ApoB100 secretion [335]. Treatment with 25-hydroxycholesterol (which inhibits proteolytic processing of SREBP-1) reverses the CYP7A1-mediated increase in mSREBP-1 levels and increases the secretion of ApoB100 [22, 335]. CYP7B1 and SREBP-1 activation are mutually regulated, with SREBP-1 repressing CYP7B1 transcription in rat McA-RH7777 hepatoma cells, likely by interacting with the basal transcriptional activator Sp1 at GC-rich sequences within the proximal promoter of CYP7B1 [338].
18 PUFAs
PUFAs are considered to compete with agonistic LXR ligands for binding to the LXR ligand-binding domain, thereby preventing the LXR/RXR heterodimer from interacting with the SREBP-1c promoter and inhibiting SREBP-1 expression [100]. In addition, independent of LXR, PUFAs suppress the proteolytic processing of SREBP-1 in mouse liver, thus disrupting the autoregulatory feedback loop by which SREBP-1 induces its own expression [99]. PUFAs also accelerate SREBP-1 mRNA degradation and, as shown for docosahexaenoic acid, reduce mSREBP-1 stability and nuclear availability through ERK- and 26S proteasome–dependent pathways, among others, in hepatocytes [101, 340].
Negative regulation of SREBP-1 by PUFAs is critical for hepatic lipid homeostasis, as exemplified by the deletion of ELOVL5 in mice [25]. ELOVL5 is involved in the conversion of essential fatty acid precursors (i.e., linoleic acid, linolenic acid) to arachidonic acid, docosapentaenoic acid, and docosahexaenoic acid, and its knockout reduces the hepatic availability of these PUFAs [341]. As a result, SREBP-1c is activated, triggering the expression of lipogenic target genes and inducing hepatic steatosis [25]. The authors propose a feedback loop whereby PUFA biosynthesis, enhanced by SREBP-1, interferes with the activation of this transcription factor [25]. A population-based investigation supports that ω3 PUFA intake through fish consumption or ω3-fatty acid supplementation reduces the risk of HCC [342]. Dietary ω6 PUFAs, by contrast, show a dose-dependent, positive association with HCC risk [343]. However, it remains unclear whether PUFAs suppress SREBP-1 activation in the human liver and whether this mechanism contributes to the proposed anti-HCC activity of ω3-fatty acids.
In HepG2 cells treated with the LXR agonist T0901317, oxidized PUFAs (i.e., the oxidized ω3-fatty acid eicosapentaenoic acid) possess superior anti-lipogenic activity over non-oxidized fatty acids, and this metabolic shift correlates with decreased SREBP-1c and PGC-1β expression [344]. Further studies are needed to clarify whether this gain in inhibitory activity compensates for the lower cellular concentrations of oxidized PUFAs relative to parental levels and whether PUFA oxidation has pathophysiological relevance in HCC. Importantly, oxygenated PUFAs are precursors of structurally diverse lipid mediators that combine potent pro-inflammatory, anti-inflammatory, pro-resolving, and immunomodulatory properties with tumor- and metastasis-regulatory functions [30, 31, 345]. Peroxidation of phospholipid-bound PUFAs, on the other hand, is a hallmark of ferroptosis, the induction of which by small molecules is currently being explored as a potential strategy against (therapy-resistant) HCC [346].
Another study in human Huh-7 and SMMC-7721 hepatoma cells silenced ACSL4 [308], an isoenzyme that couples fatty acids to coenzyme A and preferentially accepts PUFAs, which can then be incorporated into triglycerides, phospholipids, or CE esters [247]. ACSL4 is an oncogenic marker of the α-fetoprotein-high subtype of HCC and promotes HCC tumor formation and metastasis in Huh-7-grafted mice [308]. The tumor-promoting activity is partially dependent on SREBP-1, as demonstrated by SREBP-1 overexpression in ACSL4-silenced tumors. ACSL4 enhances SREBP-1 expression by stabilizing c-Myc [311], which binds directly to the SREBP-1 promoter region and activates transcription [308]. In consequence, the expression of the SREBP-1 target genes is induced and triglycerides and cholesterol accumulate. Mechanistically, ACSL4 attenuates proteasomal degradation of c-Myc in an ERK- and FBW7-dependent manner [311]. How ACSL4 modulates the ERK-FBW7-c-Myc axis is not readily understood, though the mechanism likely involves changes in the availability of PUFAs or PUFA-derived metabolites.
Complementary mechanistic insights come from the genetic manipulation of lysophosphatidylcholine acyltransferase 3 (LPCAT3) in primary mouse hepatocytes and liver [347]. The LXR target gene LPCAT3 uses PUFA-CoA (derived from ACSL4) as substrate and incorporates the acyl-chain into phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS) [348, 349]. Accordingly, LPCAT3 deletion decreases the proportion of PUFA-containing PC in the ER, reduces SREBP-1 processing to the mature nuclear form, and thereby suppresses lipogenic responses [349]. (i) Feeding, (ii) delivery of exogenous PUFA-containing PC to the ER, and (iii) LXR activation have opposite effects [349]. Dynamic changes in the PUFA composition of the ER membrane influence either the SCAP/SREBP-1 interaction or the transport of SREBP-1 to the Golgi [348]. The detailed mechanisms remain elusive and might involve phospholipid classes other than PC. In support of this hypothesis, PE with saturated fatty acids impairs the processing of the Drosophila SREBP ortholog in S2 cells [350], and inhibition of de novo phospholipid biosynthesis leads to a mislocalization of S1P and S1P to the ER (instead of the Golgi), allowing efficient processing of SREBP-1 [351]. Interestingly, supplementation of PUFA-containing PC and depletion of LPCAT3 have also been reported to decrease Akt phosphorylation by reducing the kinase’s affinity to phosphoinositides at membranes [352, 353]. Whether the associated decrease in long-term Akt signaling affects SREBP-1 expression has not been addressed.
19 Other SREBP-1 regulatory factors and mechanisms
In the following, we summarize diverse mechanisms that have been shown to regulate SREBP-1 in HCC and discuss their (pre-)clinical implications.
The multidomain adaptor protein β2-spectrin (SPTBN1), which is involved in TGFβ-SMAD3 (mothers against decapentaplegic homolog 3) signaling, among others, increases SREBP-1 activity, lipogenesis, and HCC progression in mice fed a high-fat diet or a Western diet [95]. The authors propose that caspase 3 cleaves SPTBN1 and SREBP-1 and that the N-terminal product of SPTBN1 (N-SPTBN1) interacts with cleaved SREBP-1 to stabilize the nuclear form of SREBP-1, thereby inducing the expression of target genes. Accordingly, liver-specific deletion of SPTBN1 (which is overexpressed in NASH along with caspase 3) protects mice against hepatic steatosis, fibrosis, inflammation, tissue damage, and HCC [95].
MicroRNA-27a (miR-27a) modulates cancer cell proliferation, apoptosis, migration, and invasion as well as angiogenesis and therapy resistance and has both oncogenic and tumor suppressor functions [354,355,356,357,358]. miR-27a is also preferentially expressed in HCV-infected liver and has in this context been found to repress SREBP-1 expression along with other major lipogenic regulators and enzymes, including RXRα, PPARα, PPARγ, and FASN in human HUH-7.5 hepatocellular carcinoma cells [359]. Repression of miR-27a increases cellular lipid accumulation and HCC infectivity, whereas overexpression has the opposite effect.
The related RNA binding proteins Lin28A and Lin28B (Lin28A/B) are upregulated in HCC and other cancers [360]. They initiate the post-transcriptional repression of the let-7 microRNA family, but also bind various mRNAs with roles in cell metabolism, cell cycle progression, and survival [361,362,363,364,365]. Among these mRNAs are those of SREBP-1 and SCAP, which are positively regulated by Lin28A/B in HCC cell lines [43]. By interacting with SREBP-1 and SCAP mRNA, the proto-oncogenes Lin28A/B enhance the translation and processing of SREBP-1 and stimulate tumor growth in mouse xenografts of human PLC hepatocellular carcinoma cells [43]. Mechanistically, the authors (i) demonstrate that SREBP-1, the SREBP-1 target gene SCD1, and ER stress (or the associated UPR) are involved in HCC progression; (ii) point to the imbalance of fatty acid unsaturation upon Lin28A/B silencing; and (iii) propose that Lin28A/B protects HCC cells from ER lipotoxicity [43]. Whether the recently discovered SCD1-derived lipokine PI(18:1/18:1) is enriched upon SCD1 upregulation and contributes to the tumor-protective activity of Lin28A/B has not been addressed [366].
HDGF has a highly conserved N-terminal PWWP domain and exerts oncogenic activity (related to transformation, survival, metastasis, and angiogenesis) through incompletely understood mechanisms [367,368,369,370,371]. Nuclear-localized HDGF was recently shown to bind to and act as a co-activator of SREBP-1 and to enhance lipogenic gene expression in HepG2 cells by attenuating the recruitment of the transcription repressor C-terminal binding protein 1 (CTBP1) [89]. In support of a functional role of (nuclear) HDGF in regulating SREBP-1 levels, the combined expression of the two factors indicates a poor prognosis in HCC [89]. Sequence variability exists in PWWP, and the A-type variant (in contrast to wildtype HDGF) recruits CTBP1, suppresses lipid biosynthesis, and decreases proliferation of HepG2 cells, both in vitro and in murine xenografts [89].
The transcription factor p53, a major tumor suppressor, and ferredoxin reductase (FDXR), which is involved in steroid biosynthesis, negatively regulate the maturation of SREBP-1/2 and thus keep cellular levels of triglycerides and cholesterol in check, as shown for mouse embryonic fibroblasts (MEFs), HepG2 cells, and/or mouse liver [372]. The authors further suggest, based on correlative data, that the availability of the cholesterol efflux pump ABCA1, which is induced by either p53 [373] or FDXR [372], determines SREBP-1/2 activation [372]. Deletion of p53 or FDXR (as well as the double KO) induces hepatic steatosis, inflammation, and spontaneous tumorigenesis in mice, which is, however, not limited to HCC [372]. On the other hand, p53 binds to the SREBP-1 promoter and induces SREBP-1 transcriptional activity in HepG2 cells [46]. NAD(P)H quinone oxidoreductase-1 (NQO1), which is highly expressed in HCC and associated with poor outcome, induces SREBP-1 transcription in HepG2 cells through this mechanism, specifically by preventing ubiquitination and proteasomal degradation of p53 [46]. Notably, the NQO1-p53-SREBP-1-axis stabilizes the EMT transcription factor Snail, thereby inducing lipid anabolism and EMT of HCC cells, which promotes the progression and metastasis of HCC [46].
20 Inhibitors of SREBP-1 signaling with diverse mechanisms
A large number of small molecules and some oligonucleotide-based approaches have been reported to interfere with SREBP-1 signaling in HCC (respectively HCC-promoting liver pathologies) by different, only partially understood mechanisms, as summarized in Table 1 and discussed below for selected compounds.
1-(4-Bromophenyl)-3-(pyridin-3-yl)urea (SI-1), an inhibitor of SREBP-1 activation, decreases the mRNA expression of SREBP-1 target genes in HCC cells more potently than betulin or fatostatin (FASN: IC50 = 0.3, 1.6, and 1.0 μM, respectively), lowers aerobic glycolysis, and potentiates the anti-tumoral efficacy of radiofrequency ablation towards xenograft HCC (at 0.5–5 mg/kg; peroral administration, p.o.) [44].
SREBP decoy oligodeoxynucleotides are short, double-stranded DNA sequences that mimic SREs and compete with them for binding to SREBP without generating a functional response, thereby blocking the expression of SREBP target genes [374]. They have been shown to inhibit the expression of SREBP-1c, ACC1, FASN, SCD1, and HMGCR in hyperlipidemic mice fed a high-fat diet, thereby alleviating the associated inflammation as indicated by the reduction in pro-inflammatory cytokine levels [115].
Scientists at Merck developed siRNA oligonucleotide-lipid nanoparticles (siRNA-LNPs) targeting SCAP and demonstrated that this approach is effective in reducing hepatic SCAP mRNA expression [119]. As a result, hyperlipidemia is attenuated in rhesus monkeys and mice [119, 120], in the latter accompanied by decreased hepatic Ldlr and proprotein convertase subtilisin kexin/type 9 (Pcsk9) expression, repression of Srebp-regulated genes, and inhibition of de novo lipogenesis [119]. The serine kinase PCSK9, which is a therapeutic target for lipid-lowering drugs, is secreted by hepatocytes and subjects LDL receptors to lysosomal degradation [375].
The diarylthiazole fatostatin interacts with SCAP, inhibits its glycosylation, and blocks the transport of SREBP-1 from the ER to the Golgi, thereby suppressing SREBP-1 maturation [117, 376]. Fatostatin attenuates hepatic steatosis in obese mice while reducing body weight and blood glucose levels [117] and exhibits anti-tumoral/growth-retarding [377,378,379] and ferroptosis-inducing activity [379], which has been ascribed to impaired SREBP-1 activation [376]. We expect that fatostatin may also be cytotoxic to HCC cells, although this has not been explicitly demonstrated.
The pentacyclic lupane-type triterpenoid betulin from birch bark has a broad spectrum of pharmacological activities, among others, directed against metabolic disorders and cancer, including HCC [38, 66, 380]. Pleiotropic anti-tumoral mechanisms have been proposed for betulin, including inhibition of SREBP-1/2 maturation, which reduces fatty acid and cholesterol biosynthesis in HCC cells (2.3–100 μM betulin) and DEN-induced HCC in mice (gavage of 100 mg betulin/kg/day) [38, 66, 116]. Mechanistically, betulin physically interacts with SCAP (at 100 μM) and thereby promotes the interaction of SCAP with Insig1/2 to retain SREBPs at the ER [116]. Suppression of SREBP-1/2 and SREBP target genes in experimental murine HCC was accompanied by decreased mRNA expression of pro-inflammatory cytokines, such as TNFα [38]. While the authors confirm a functional link between SREBP processing and pro-inflammatory cytokine expression, the cytokine-lowering mechanism of betulin and its contribution to the anti-HCC activity remain diffuse, especially when considering that betulin may also target the TLR4 and nuclear factor (NF)-κB pathways [39].
Sulforaphane, an isothiocyanate from cruciferous vegetables, and the desaturated analog sulforaphene (30–100 μM, each) inhibit lipogenic enzyme expression in human Huh-7 hepatoma cells by promoting the ubiquitination and proteasomal degradation of pSREBP-1/2 in a SCAP-independent manner [122]. Central to the SREBP-1a-degrading activity of sulforaphane and sulforaphene is the SREPB-1a amino acid sequence from 595 to 784. The direct molecular target of sulforaphane responsible for the induction of SREBP-1/2 degradation remains elusive. Neither does sulforaphane interact directly with SREBPs, nor are known targets of sulforaphane, i.e., Kelch-like ECH-associated protein 1/nuclear factor erythroid 2-related factor 2 (KEAP1/NRF2) and heat shock protein 27 (HSP27), involved in pSREBP degradation [122]. Another study proposed that sulforaphane (1–20 μM; 5–20 mg/kg/day, p.o.) reduces hepatic lipogenic gene expression in rats on a high-fat diet by repressing the ER stress sensor protein kinase-like ER kinase (PERK) and decreasing SREPB-1 expression [123].
The bufadienolide cinobufotalin (0.1–0.4 μM) from the skin secretion of the giant toad inhibits both SREBP-1 expression and the binding of SREBP-1 to SREs in HepG2 cells, seemingly by interacting directly with the transcription factor, which together markedly represses the expression of lipogenic enzymes [61]. Cinobufotalin (2.5–5 mg/kg, i.p.) is effective in vivo and reduces lipogenesis and HCC tumor growth in grafted mice, the latter likely by inducing G2/M cell cycle arrest and apoptosis [61]. A meta-analysis of 27 clinical trials involving 2079 advanced HCC patients indicates that the combination of hepatic arterial chemoembolization (TACE) with adjuvant cinobufotalin injection is safe and more effective than TACE alone for the treatment of end-stage HCC [381].
Diverse small molecules and complex mixtures have been shown to decrease SREBP-1 expression or maturation in HCC cell lines, though the mode of action has often remained elusive (Table 1). These include pyridine co-ligand functionalized Pt(II) complexes [190]; the retinoic acid receptor β2 agonists AC261066 and AC55649 [176]; GPR40 agonists, such as GW9508, AMG-1638, and docosahexaenoic acid [179, 180]; and ethanolic extracts of several herbs, such as Zhiheshouwu (Polygoni multiflori Radix Praeparata) and Liriope platyphylla root [382, 383].
21 Conclusion and perspective
Malignant transformation to HCC induces SREBP-1 signaling through a broad spectrum of regulatory mechanisms that enhance SREBP-1 expression, maturation, protein stability, and nuclear activity. This metabolic reprogramming confers advantages in survival, growth, proliferation, and dissemination to HCC, but also renders tumors sensitive to anti-lipogenic treatment. While the mechanistic insights into SREBP-1 regulation and function are rapidly increasing, pharmacological strategies that selectively target SREBP-1 signaling are still in their infancy. The main reasons for this are the limited availability of high-throughput screening assays for SREBP-1-interacting small molecules, the lack of obvious ligand binding pockets, and incomplete structural information. Crystal structures of SREBP-1 that could aid in the rational design of respective ligands have been solved, but are limited to the yeast ortholog and the N-terminal bHLH-Zip domain of the human transcription factor [384,385,386].
The vast majority of agents targeting SREBP-1 signaling are non-selective, either because of polypharmacological activities or because the upstream targets regulate multiple signaling pathways in addition to SREBP-1 activation/induction. Furthermore, many pathways, including kinase cascades (e.g., PI3K-Akt, mTORC1, AMPK) and transcription factors/co-activators (e.g., LXR), indirectly regulate SREBP-1 signaling. Because these pathways regulate multiple other cellular processes besides SREBP-1 that are involved in tumorigenesis, it is difficult to assess from genetic manipulation studies the extent to which the interference with SREBP-1 contributes to HCC suppression. Thus, many of the agents listed in Table 1 have been evaluated in pre-clinical studies and some in clinical trials, with the effective doses (in mg/kg body weight) used in (pre-)clinical studies reported whenever available. For example, the pan-kinase inhibitor sorafenib is a first-line treatment for HCC [387, 388], and the red wine stilbene resveratrol has been the subject of numerous clinical and epidemiological studies and meta-analyses (including a phase I trial in patients with liver metastases) [389]. AMPK activators have also been intensively studied in recent years for the treatment of metabolic diseases, including cancer, and several compounds have entered clinical evaluation [353]. While these compounds have been reported to interfere with SREBP-1 signaling and it is likely that this effect contributes to their overall efficacy, clinical and pre-clinical data do not allow conclusions as to whether the interference with SREBP-1 signaling mediates the observed beneficial and adverse effects.
Results from pharmacological approaches are even more difficult to interpret when considering that the vast majority of SREBP-1 modulators have been shown to have off-target effects; many other compounds have not been adequately studied. Available selective approaches to inhibit SREBP-1 signaling are largely limited to (i) the interaction of SREBP-1 and the upstream kinase PKM2 via peptide ligands (P8) [181], (ii) an apparent direct interaction of cinobufotalin with SREBP-1 (although little is known about potential other targets for this traditional Chinese medicine) [61], and (iii) oligonucleotide-based approaches via siRNA or decoy oligonucleotides [115, 119, 120]. Biopharmaceuticals such as therapeutic antibodies or SREBP-1-interacting proteins, which are expected to provide superior SREBP-1 selectivity, have not been explored so far. Of these more selective approaches, only cinobufotalin has been tested and shown to be effective in an in vivo HCC model [61] and clinical trials [381], while oligonucleotide approaches have been studied in hyperlipidemic animals (where they reduced lipogenesis and hepatic LDL uptake) [119, 120], and knowledge of P8 is limited to cell-based studies [181]. Such strategies may have the potential to achieve selectivity for SREBP-1 targeting. Other promising strategies to narrow down putative side effects (which we have not or only partially addressed here) include (i) selective inhibition of SREBP-1 target genes, such as ACC, FASN, SCD1, and ACLY [32, 390,391,392]; (ii) interference with defined, context-specific SREBP-1 regulators, e.g., PKD3 or CRTC2 [128, 198, 204]; (iii) functionalized nanoparticles for HCC targeting [393,394,395]; and (iv) tumor-specific gene therapy [223, 396].
Based on the above, the (still incompletely pharmacologically characterized) cinobufotalin seems to be currently at the forefront of agents that target SREBP-1 signaling with some selectivity. Nanoparticle-based approaches that deliver SREBP-1-interacting peptides or siRNA to target SREBP-1, SCAP, or other signaling molecules, as well as (not yet explored) therapeutic antibodies directed against these factors, hold great promise for the future. On the other hand, it is questionable whether selectivity is actually desirable to achieve efficacy against a complex disease such as HCC. Therefore, polypharmacological approaches based on the rational inhibition of SREBP-1, as already realized in several drugs and drug candidates, may pave the way to an improved clinical efficacy of rationally designed anti-cancer drugs, particularly in the treatment of HCC.
In addition, interesting new links between SREBP-1 and resistance to therapy (including chemo- and radioresistance) are emerging, driven in part by multiomics approaches [397,398,399,400], but the number of such studies exploring the role of SREBP-1 in therapy-resistant HCC is still very limited [27, 37, 44, 66, 71,72,73,74], and further research in this area is urgently needed. In this context, the bivalent role of SREBP-1 in conveying resistance in cancer (HCC) should be mentioned. High SREBP-1 levels increase HCC aggressiveness and resistance to classical chemotherapeutics, while sensitizing tumors to anti-lipogenic strategies and alternative forms of programmed cell death, such as ferroptosis [27, 42, 74, 98].
In summary, selective SREBP-1 inhibitors are in high demand to investigate the pleiotropic, context-dependent functions of SREBP-1. Current anti-HCC strategies are instead dominated by multitarget small molecules that exhibit (subordinate) SREBP-1 inhibition, broadly interfere with lipogenesis, and may target additional cancer-promoting pathways. Whether they are inferior or superior towards monopharmacological approaches in terms of efficacy and safety remains to be determined in future studies.
Data availability
This review article does not contain original data but summarizes and discusses published data.
References
European Association For The Study Of The Liver. (2018). EASL clinical practice guidelines: Management of hepatocellular carcinoma. Journal of Hepatology, 69(1), 182–236. https://doi.org/10.1016/j.jhep.2018.03.019
Vogel, A., Meyer, T., Sapisochin, G., Salem, R., & Saborowski, A. (2022). Hepatocellular carcinoma. The Lancet, 400(10360), 1345–1362. https://doi.org/10.1016/S0140-6736(22)01200-4
Tilg, H., Adolph, T. E., Dudek, M., & Knolle, P. (2021). Non-alcoholic fatty liver disease: The interplay between metabolism, microbes and immunity. Nature Metabolism, 3(12), 1596–1607. https://doi.org/10.1038/s42255-021-00501-9
Muir, K., Hazim, A., He, Y., Peyressatre, M., Kim, D. Y., Song, X., & Beretta, L. (2013). Proteomic and lipidomic signatures of lipid metabolism in NASH-associated hepatocellular carcinoma. Cancer Research, 73(15), 4722–4731. https://doi.org/10.1158/0008-5472.CAN-12-3797
Wang, M., Han, J., Xing, H., Zhang, H., Li, Z., Liang, L., Li, C., Dai, S., Wu, M., Shen, F., & Yang, T. (2016). Dysregulated fatty acid metabolism in hepatocellular carcinoma. Hepatic Oncology, 3(4), 241–251. https://doi.org/10.2217/hep-2016-0012
Calvisi, D. F., Wang, C., Ho, C., Ladu, S., Lee, S. A., Mattu, S., Destefanis, G., Delogu, S., Zimmermann, A., Ericsson, J., Brozzetti, S., Staniscia, T., Chen, X., Dombrowski, F., & Evert, M. (2011). Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology, 140(3), 1071–1083. https://doi.org/10.1053/j.gastro.2010.12.006
Snaebjornsson, M. T., Janaki-Raman, S., & Schulze, A. (2020). Greasing the wheels of the cancer machine: The role of lipid metabolism in cancer. Cell Metabolism, 31(1), 62–76. https://doi.org/10.1016/j.cmet.2019.11.010
Li, T., Weng, J., Zhang, Y., Liang, K., Fu, G., Li, Y., Bai, X., & Gao, Y. (2019). mTOR direct crosstalk with STAT5 promotes de novo lipid synthesis and induces hepatocellular carcinoma. Cell Death & Disease, 10(8), 619. https://doi.org/10.1038/s41419-019-1828-2
Li, C., Yang, W., Zhang, J., Zheng, X., Yao, Y., Tu, K., & Liu, Q. (2014). SREBP-1 has a prognostic role and contributes to invasion and metastasis in human hepatocellular carcinoma. International Journal of Molecular Sciences, 15(5), 7124–7138. https://doi.org/10.3390/ijms15057124
Shimano, H. (2001). Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Progress in Lipid Research, 40(6), 439–452. https://doi.org/10.1016/S0163-7827(01)00010-8
Jeon, Y. G., Kim, Y. Y., Lee, G., & Kim, J. B. (2023). Physiological and pathological roles of lipogenesis. Nature Metabolism, 5(5), 735–759. https://doi.org/10.1038/s42255-023-00786-y
Soyal, S. M., Nofziger, C., Dossena, S., Paulmichl, M., & Patsch, W. (2015). Targeting SREBPs for treatment of the metabolic syndrome. Trends in Pharmacological Sciences, 36(6), 406–416. https://doi.org/10.1016/j.tips.2015.04.010
Zhao, Q., Lin, X., & Wang, G. (2022). Targeting SREBP-1-mediated lipogenesis as potential strategies for cancer. Frontiers in Oncology, 12, 952371. https://doi.org/10.3389/fonc.2022.952371
Athanikar, J. N., & Osborne, T. F. (1998). Specificity in cholesterol regulation of gene expression by coevolution of sterol regulatory DNA element and its binding protein. Proceedings of the National Academy of Sciences of the United States of America, 95(9), 4935–4940. https://doi.org/10.1073/pnas.95.9.4935
Amemiya-Kudo, M., Shimano, H., Yoshikawa, T., Yahagi, N., Hasty, A. H., Okazaki, H., Tamura, Y., Shionoiri, F., Iizuka, Y., Ohashi, K., Osuga, J., Harada, K., Gotoda, T., Sato, R., Kimura, S., Ishibashi, S., & Yamada, N. (2000). Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. Journal of Biological Chemistry, 275(40), 31078–31085. https://doi.org/10.1074/jbc.M005353200
Horton, J. D., Goldstein, J. L., & Brown, M. S. (2002). SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. The Journal of Clinical Investigation, 109(9), 1125–1131. https://doi.org/10.1172/JCI15593
Shimano, H., Horton, J. D., Shimomura, I., Hammer, R. E., Brown, M. S., & Goldstein, J. L. (1997). Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. The Journal of Clinical Investigation, 99(5), 846–854. https://doi.org/10.1172/JCI119248
Foretz, M., Guichard, C., Ferre, P., & Foufelle, F. (1999). Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proceedings of the National Academy of Sciences of the United States of America, 96(22), 12737–12742. https://doi.org/10.1073/pnas.96.22.12737
Shechter, I., Dai, P., Huo, L., & Guan, G. (2003). IDH1 gene transcription is sterol regulated and activated by SREBP-1a and SREBP-2 in human hepatoma HepG2 cells: Evidence that IDH1 may regulate lipogenesis in hepatic cells. Journal of Lipid Research, 44(11), 2169–2180. https://doi.org/10.1194/jlr.M300285-JLR200
Lee, J. N., Song, B., DeBose-Boyd, R. A., & Ye, J. (2006). Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. Journal of Biological Chemistry, 281(51), 39308–39315. https://doi.org/10.1074/jbc.M608999200
Liu, T. F., Tang, J. J., Li, P. S., Shen, Y., Li, J. G., Miao, H. H., Li, B. L., & Song, B. L. (2012). Ablation of gp78 in liver improves hyperlipidemia and insulin resistance by inhibiting SREBP to decrease lipid biosynthesis. Cell Metabolism, 16(2), 213–225. https://doi.org/10.1016/j.cmet.2012.06.014
Wang, X., Sato, R., Brown, M. S., Hua, X., & Goldstein, J. L. (1994). SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell, 77(1), 53–62. https://doi.org/10.1016/0092-8674(94)90234-8
Radhakrishnan, A., Ikeda, Y., Kwon, H. J., Brown, M. S., & Goldstein, J. L. (2007). Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Oxysterols block transport by binding to Insig. Proceedings of the National Academy of Sciences of the United States of America, 104(16), 6511–6518. https://doi.org/10.1073/pnas.0700899104
Adams, C. M., Reitz, J., De Brabander, J. K., Feramisco, J. D., Li, L., Brown, M. S., & Goldstein, J. L. (2004). Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. The Journal of Biological Chemistry, 279(50), 52772–52780. https://doi.org/10.1074/jbc.M410302200
Moon, Y. A., Hammer, R. E., & Horton, J. D. (2009). Deletion of ELOVL5 leads to fatty liver through activation of SREBP-1c in mice. Journal of Lipid Research, 50(3), 412–423. https://doi.org/10.1194/jlr.M800383-JLR200
Williams, K. J., Argus, J. P., Zhu, Y., Wilks, M. Q., Marbois, B. N., York, A. G., Kidani, Y., Pourzia, A. L., Akhavan, D., Lisiero, D. N., Komisopoulou, E., Henkin, A. H., Soto, H., Chamberlain, B. T., Vergnes, L., Jung, M. E., Torres, J. Z., Liau, L. M., Christofk, H. R., et al. (2013). An essential requirement for the SCAP/SREBP signaling axis to protect cancer cells from lipotoxicity. Cancer Research, 73(9), 2850–2862. https://doi.org/10.1158/0008-5472.CAN-13-0382-T
Liang, D., Minikes, A. M., & Jiang, X. (2022). Ferroptosis at the intersection of lipid metabolism and cellular signaling. Molecular Cell, 82(12), 2215–2227. https://doi.org/10.1016/j.molcel.2022.03.022
Zhang, M., Wei, T., Zhang, X., & Guo, D. (2022). Targeting lipid metabolism reprogramming of immunocytes in response to the tumor microenvironment stressor: A potential approach for tumor therapy. Frontiers in Immunology, 13, 937406. https://doi.org/10.3389/fimmu.2022.937406
Park, H. Y., Kang, H. S., & Im, S. S. (2018). Recent insight into the correlation of SREBP-mediated lipid metabolism and innate immune response. The Journal of Molecular Endocrinology, 61(3), R123–r131. https://doi.org/10.1530/jme-17-0289
Dyall, S. C., Balas, L., Bazan, N. G., Brenna, J. T., Chiang, N., da Costa Souza, F., Dalli, J., Durand, T., Galano, J. M., Lein, P. J., Serhan, C. N., & Taha, A. Y. (2022). Polyunsaturated fatty acids and fatty acid-derived lipid mediators: Recent advances in the understanding of their biosynthesis, structures, and functions. Progress in Lipid Research, 86, 101165. https://doi.org/10.1016/j.plipres.2022.101165
Shimizu, T. (2009). Lipid mediators in health and disease: Enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annual Review of Pharmacology and Toxicology, 49, 123–150. https://doi.org/10.1146/annurev.pharmtox.011008.145616
Cheng, C., Geng, F., Cheng, X., & Guo, D. (2018). Lipid metabolism reprogramming and its potential targets in cancer. Cancer Communications, 38(1), 27. https://doi.org/10.1186/s40880-018-0301-4
Pope, E. D., 3rd, Kimbrough, E. O., Vemireddy, L. P., Surapaneni, P. K., Copland, J. A., 3rd, & Mody, K. (2019). Aberrant lipid metabolism as a therapeutic target in liver cancer. Expert Opinion on Therapeutic Targets, 23(6), 473–483. https://doi.org/10.1080/14728222.2019.1615883
Su, F., Tang, X., Li, G., Koeberle, A., & Liu, B. (2021). SIRT7–SREBP1 restrains cancer cell metabolic reprogramming by upregulating IDH1. Genome Instability & Disease. https://doi.org/10.1007/s42764-021-00031-4
Hu, Q., Mao, Y., Liu, M., Luo, R., Jiang, R., & Guo, F. (2020). The active nuclear form of SREBP1 amplifies ER stress and autophagy via regulation of PERK. The FEBS Journal, 287(11), 2348–2366. https://doi.org/10.1111/febs.15144
Zhou, C., Qian, W., Li, J., Ma, J., Chen, X., Jiang, Z., Cheng, L., Duan, W., Wang, Z., Wu, Z., Ma, Q., & Li, X. (2019). High glucose microenvironment accelerates tumor growth via SREBP1-autophagy axis in pancreatic cancer. Journal of Experimental & Clinical Cancer Research, 38(1), 302. https://doi.org/10.1186/s13046-019-1288-7
Zhao, Y., Li, M., Yao, X., Fei, Y., Lin, Z., Li, Z., Cai, K., Zhao, Y., & Luo, Z. (2020). HCAR1/MCT1 regulates tumor ferroptosis through the lactate-mediated AMPK-SCD1 activity and its therapeutic implications. Cell Reports, 33(10), 108487. https://doi.org/10.1016/j.celrep.2020.108487
Li, N., Zhou, Z. S., Shen, Y., Xu, J., Miao, H. H., Xiong, Y., Xu, F., Li, B. L., Luo, J., & Song, B. L. (2017). Inhibition of the sterol regulatory element-binding protein pathway suppresses hepatocellular carcinoma by repressing inflammation in mice. Hepatology, 65(6), 1936–1947. https://doi.org/10.1002/hep.29018
Oishi, Y., Spann, N. J., Link, V. M., Muse, E. D., Strid, T., Edillor, C., Kolar, M. J., Matsuzaka, T., Hayakawa, S., Tao, J., Kaikkonen, M. U., Carlin, A. F., Lam, M. T., Manabe, I., Shimano, H., Saghatelian, A., & Glass, C. K. (2017). SREBP1 contributes to resolution of pro-inflammatory TLR4 Signaling By Reprogramming Fatty Acid Metabolism. Cell Metabolism, 25(2), 412–427. https://doi.org/10.1016/j.cmet.2016.11.009
Chyau, C. C., Wang, H. F., Zhang, W. J., Chen, C. C., Huang, S. H., Chang, C. C., & Peng, R. Y. (2020). Antrodan alleviates high-fat and high-fructose diet-induced fatty liver disease in C57BL/6 mice model via AMPK/Sirt1/SREBP-1c/PPARγ pathway. International Journal of Molecular Sciences, 21(1). https://doi.org/10.3390/ijms21010360
Guri, Y., Colombi, M., Dazert, E., Hindupur, S. K., Roszik, J., Moes, S., Jenoe, P., Heim, M. H., Riezman, I., Riezman, H., & Hall, M. N. (2017). mTORC2 promotes tumorigenesis via lipid synthesis. Cancer Cell, 32(6), 807–823 e812. https://doi.org/10.1016/j.ccell.2017.11.011
Stockwell, B. R. (2022). Ferroptosis turns 10: Emerging mechanisms, physiological functions, and therapeutic applications. Cell, 185(14), 2401–2421. https://doi.org/10.1016/j.cell.2022.06.003
Zhang, Y., Li, C., Hu, C., Wu, Q., Cai, Y., Xing, S., Lu, H., Wang, L. H., Sun, L., Li, T., He, X., Zhong, X., Wang, J., Gao, P., Smith, Z. J., Jia, W., & Zhang, H. (2019). Lin28 enhances de novo fatty acid synthesis to promote cancer progression via SREBP-1. EMBO Reports, 20(10), e48115. https://doi.org/10.15252/embr.201948115
Zou, X. Z., Hao, J. F., & Zhou, X. H. (2021). Inhibition of SREBP-1 activation by a novel small-molecule inhibitor enhances the sensitivity of hepatocellular carcinoma tissue to radiofrequency ablation. Frontiers in Oncology, 11, 796152. https://doi.org/10.3389/fonc.2021.796152
Husain, A., Chiu, Y. T., Sze, K. M., Ho, D. W., Tsui, Y. M., Suarez, E. M. S., Zhang, V. X., Chan, L. K., Lee, E., Lee, J. M., Cheung, T. T., Wong, C. C., Chung, C. Y., & Ng, I. O. (2022). Ephrin-A3/EphA2 axis regulates cellular metabolic plasticity to enhance cancer stemness in hypoxic hepatocellular carcinoma. Journal of Hepatology, 77(2), 383–396. https://doi.org/10.1016/j.jhep.2022.02.018
Wang, X., Liu, Y., Han, A., Tang, C., Xu, R., Feng, L., Yang, Y., Chen, L., & Lin, Z. (2022). The NQO1/p53/SREBP1 axis promotes hepatocellular carcinoma progression and metastasis by regulating Snail stability. Oncogene. https://doi.org/10.1038/s41388-022-02477-6
Bhalla, K., Hwang, B. J., Dewi, R. E., Twaddel, W., Goloubeva, O. G., Wong, K. K., Saxena, N. K., Biswal, S., & Girnun, G. D. (2012). Metformin prevents liver tumorigenesis by inhibiting pathways driving hepatic lipogenesis. Cancer Prevention Research, 5(4), 544–552. https://doi.org/10.1158/1940-6207.CAPR-11-0228
Ma, A. P. Y., Yeung, C. L. S., Tey, S. K., Mao, X., Wong, S. W. K., Ng, T. H., Ko, F. C. F., Kwong, E. M. L., Tang, A. H. N., Ng, I. O., Cai, S. H., Yun, J. P., & Yam, J. W. P. (2021). Suppression of ACADM-mediated fatty acid oxidation promotes hepatocellular carcinoma via aberrant CAV1/SREBP1 signaling. Cancer Research, 81(13), 3679–3692. https://doi.org/10.1158/0008-5472.CAN-20-3944
Guo, D., Bell, E. H., Mischel, P., & Chakravarti, A. (2014). Targeting SREBP-1-driven lipid metabolism to treat cancer. Current pharmaceutical design, 20(15), 2619–2626. https://doi.org/10.2174/13816128113199990486
Shao, W., & Espenshade, P. J. (2012). Expanding roles for SREBP in metabolism. Cell Metabolism, 16(4), 414–419. https://doi.org/10.1016/j.cmet.2012.09.002
Paul, B., Lewinska, M., & Andersen, J. B. (2022). Lipid alterations in chronic liver disease and liver cancer. JHEP Reports, 4(6), 100479. https://doi.org/10.1016/j.jhepr.2022.100479
Moslehi, A., & Hamidi-Zad, Z. (2018). Role of SREBPs in liver diseases: A mini-review. Journal of Clinical and Translational Hepatology, 6(3), 332–338. https://doi.org/10.14218/jcth.2017.00061
Shimano, H., & Sato, R. (2017). SREBP-regulated lipid metabolism: Convergent physiology - divergent pathophysiology. Nature Reviews Endocrinology, 13(12), 710–730. https://doi.org/10.1038/nrendo.2017.91
Wang, Y., Zhang, Y., Wang, Z., Yu, L., Chen, K., Xie, Y., Liu, Y., Liang, W., Zheng, Y., Zhan, Y., & Ding, Y. (2022). The interplay of transcriptional coregulator NUPR1 with SREBP1 promotes hepatocellular carcinoma progression via upregulation of lipogenesis. Cell Death Discovery, 8(1), 431. https://doi.org/10.1038/s41420-022-01213-z
Zhao, Z., Zhong, L., He, K., Qiu, C., Li, Z., Zhao, L., & Gong, J. (2019). Cholesterol attenuated the progression of DEN-induced hepatocellular carcinoma via inhibiting SCAP mediated fatty acid de novo synthesis. Biochemical and Biophysical Research Communications, 509(4), 855–861. https://doi.org/10.1016/j.bbrc.2018.12.181
Yin, F., Sharen, G., Yuan, F., Peng, Y., Chen, R., Zhou, X., Wei, H., Li, B., Jing, W., & Zhao, J. (2017). TIP30 regulates lipid metabolism in hepatocellular carcinoma by regulating SREBP1 through the Akt/mTOR signaling pathway. Oncogenesis, 6(6), e347. https://doi.org/10.1038/oncsis.2017.49
Liu, Y., Ren, H., Zhou, Y., Shang, L., Zhang, Y., Yang, F., & Shi, X. (2019). The hypoxia conditioned mesenchymal stem cells promote hepatocellular carcinoma progression through YAP mediated lipogenesis reprogramming. Journal of Experimental & Clinical Cancer Research, 38(1), 228. https://doi.org/10.1186/s13046-019-1219-7
Pattanayak, S. P., Bose, P., Sunita, P., Siddique, M. U. M., & Lapenna, A. (2018). Bergapten inhibits liver carcinogenesis by modulating LXR/PI3K/Akt and IDOL/LDLR pathways. Biomedicine & Pharmacotherapy, 108, 297–308. https://doi.org/10.1016/j.biopha.2018.08.145
Gouw, A. M., Margulis, K., Liu, N. S., Raman, S. J., Mancuso, A., Toal, G. G., Tong, L., Mosley, A., Hsieh, A. L., Sullivan, D. K., Stine, Z. E., Altman, B. J., Schulze, A., Dang, C. V., Zare, R. N., & Felsher, D. W. (2019). The MYC oncogene cooperates with sterol-regulated element-binding protein to regulate lipogenesis essential for neoplastic growth. Cell Metabolism, 30(3), 556–572.e555. https://doi.org/10.1016/j.cmet.2019.07.012
Yamashita, T., Honda, M., Takatori, H., Nishino, R., Minato, H., Takamura, H., Ohta, T., & Kaneko, S. (2009). Activation of lipogenic pathway correlates with cell proliferation and poor prognosis in hepatocellular carcinoma. Journal of Hepatology, 50(1), 100–110. https://doi.org/10.1016/j.jhep.2008.07.036
Meng, H., Shen, M., Li, J., Zhang, R., Li, X., Zhao, L., Huang, G., & Liu, J. (2021). Novel SREBP1 inhibitor cinobufotalin suppresses proliferation of hepatocellular carcinoma by targeting lipogenesis. European Journal of Pharmacology, 906, 174280. https://doi.org/10.1016/j.ejphar.2021.174280
Yang, N., Li, C., Li, H., Liu, M., Cai, X., Cao, F., Feng, Y., Li, M., & Wang, X. (2019). Emodin induced SREBP1-dependent and SREBP1-independent apoptosis in hepatocellular carcinoma cells. Frontiers in Pharmacology, 10, 709. https://doi.org/10.3389/fphar.2019.00709
Tian, Y., Wong, V. W., Wong, G. L., Yang, W., Sun, H., Shen, J., Tong, J. H., Go, M. Y., Cheung, Y. S., Lai, P. B., Zhou, M., Xu, G., Huang, T. H., Yu, J., To, K. F., Cheng, A. S., & Chan, H. L. (2015). Histone deacetylase HDAC8 promotes insulin resistance and β-catenin activation in NAFLD-associated hepatocellular carcinoma. Cancer Research, 75(22), 4803–4816. https://doi.org/10.1158/0008-5472.Can-14-3786
Zhou, C., Qian, W., Ma, J., Cheng, L., Jiang, Z., Yan, B., Li, J., Duan, W., Sun, L., Cao, J., Wang, F., Wu, E., Wu, Z., Ma, Q., & Li, X. (2019). Resveratrol enhances the chemotherapeutic response and reverses the stemness induced by gemcitabine in pancreatic cancer cells via targeting SREBP1. Cell Proliferation, 52(1), e12514. https://doi.org/10.1111/cpr.12514
Heo, M. J., Kang, S. H., Kim, Y. S., Lee, J. M., Yu, J., Kim, H. R., Lim, H., Kim, K. M., Jung, J., Jeong, L. S., Moon, A., & Kim, S. G. (2020). UBC12-mediated SREBP-1 neddylation worsens metastatic tumor prognosis. International Journal of Cancer, 147(9), 2550–2563. https://doi.org/10.1002/ijc.33113
Yin, F., Feng, F., Wang, L., Wang, X., Li, Z., & Cao, Y. (2019). SREBP-1 inhibitor Betulin enhances the antitumor effect of Sorafenib on hepatocellular carcinoma via restricting cellular glycolytic activity. Cell Death & Disease, 10(9), 672. https://doi.org/10.1038/s41419-019-1884-7
Andrade, J. M., Paraíso, A. F., de Oliveira, M. V., Martins, A. M., Neto, J. F., Guimarães, A. L., de Paula, A. M., Qureshi, M., & Santos, S. H. (2014). Resveratrol attenuates hepatic steatosis in high-fat fed mice by decreasing lipogenesis and inflammation. Nutrition, 30(7-8), 915–919. https://doi.org/10.1016/j.nut.2013.11.016
Jeon, T. I., & Osborne, T. F. (2012). SREBPs: Metabolic integrators in physiology and metabolism. Trends in Endocrinology and Metabolism, 23(2), 65–72. https://doi.org/10.1016/j.tem.2011.10.004
Sun, H., Yang, W., Tian, Y., Zeng, X., Zhou, J., Mok, M. T. S., Tang, W., Feng, Y., Xu, L., Chan, A. W. H., Tong, J. H., Cheung, Y. S., Lai, P. B. S., Wang, H. K. S., Tsang, S. W., Chow, K. L., Hu, M., Liu, R., Huang, L., et al. (2018). An inflammatory-CCRK circuitry drives mTORC1-dependent metabolic and immunosuppressive reprogramming in obesity-associated hepatocellular carcinoma. Nature communications, 9(1), 5214. https://doi.org/10.1038/s41467-018-07402-8
Santos, P. M., Menk, A. V., Shi, J., Tsung, A., Delgoffe, G. M., & Butterfield, L. H. (2019). Tumor-derived α-fetoprotein suppresses fatty acid metabolism and oxidative phosphorylation in dendritic cells. Cancer Immunology Research, 7(6), 1001–1012. https://doi.org/10.1158/2326-6066.Cir-18-0513
Fang, Y., Zhan, Y., Xie, Y., Du, S., Chen, Y., Zeng, Z., Zhang, Y., Chen, K., Wang, Y., Liang, L., Ding, Y., & Wu, D. (2022). Integration of glucose and cardiolipin anabolism confers radiation resistance of HCC. Hepatology, 75(6), 1386–1401. https://doi.org/10.1002/hep.32177
Xu, G. L., Ni, C. F., Liang, H. S., Xu, Y. H., Wang, W. S., Shen, J., Li, M. M., & Zhu, X. L. (2020). Upregulation of PD-L1 expression promotes epithelial-to-mesenchymal transition in sorafenib-resistant hepatocellular carcinoma cells. Gastroenterology Report, 8(5), 390–398. https://doi.org/10.1093/gastro/goaa049
Xie, L., Liu, M., Cai, M., Huang, W., Guo, Y., Liang, L., Cai, W., Liu, J., Liang, W., Tan, Y., Lai, M., Lin, L., & Zhu, K. (2023). Regorafenib enhances anti-tumor efficacy of immune checkpoint inhibitor by regulating IFN-γ/NSDHL/SREBP1/TGF-β1 axis in hepatocellular carcinoma. Biomedicine & Pharmacotherapy, 159, 114254. https://doi.org/10.1016/j.biopha.2023.114254
Yi, J., Zhu, J., Wu, J., Thompson, C. B., & Jiang, X. (2020). Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proceedings of the National Academy of Sciences of the United States of America, 117(49), 31189–31197. https://doi.org/10.1073/pnas.2017152117
Engelking, L. J., Cantoria, M. J., Xu, Y., & Liang, G. (2018). Developmental and extrahepatic physiological functions of SREBP pathway genes in mice. Seminars in Cell & Developmental Biology, 81, 98–109. https://doi.org/10.1016/j.semcdb.2017.07.011
Kobayashi, M., Fujii, N., Narita, T., & Higami, Y. (2018). SREBP-1c-dependent metabolic remodeling of white adipose tissue by caloric restriction. International Journal of Molecular Sciences, 19(11). https://doi.org/10.3390/ijms19113335
Chen, G., Liang, G., Ou, J., Goldstein, J. L., & Brown, M. S. (2004). Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proceedings of the National Academy of Sciences of the United States of America, 101(31), 11245–11250. https://doi.org/10.1073/pnas.0404297101
Wang, N., Liu, Y., Ma, Y., & Wen, D. (2018). Hydroxytyrosol ameliorates insulin resistance by modulating endoplasmic reticulum stress and prevents hepatic steatosis in diet-induced obesity mice. The Journal of Nutritional Biochemistry, 57, 180–188. https://doi.org/10.1016/j.jnutbio.2018.03.018
Yan, J. B., Lai, C. C., Jhu, J. W., Gongol, B., Marin, T. L., Lin, S. C., Chiu, H. Y., Yen, C. J., Wang, L. Y., & Peng, I. C. (2020). Insulin and metformin control cell proliferation by regulating TDG-mediated DNA demethylation in liver and breast cancer cells. Molecular Therapy Oncolytics, 18, 282–294. https://doi.org/10.1016/j.omto.2020.06.010
Owen, J. L., Zhang, Y., Bae, S. H., Farooqi, M. S., Liang, G., Hammer, R. E., Goldstein, J. L., & Brown, M. S. (2012). Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proceedings of the National Academy of Sciences of the United States of America, 109(40), 16184–16189. https://doi.org/10.1073/pnas.1213343109
Costet, P., Cariou, B., Lambert, G., Lalanne, F., Lardeux, B., Jarnoux, A. L., Grefhorst, A., Staels, B., & Krempf, M. (2006). Hepatic PCSK9 expression is regulated by nutritional status via insulin and sterol regulatory element-binding protein 1c. Journal of Biological Chemistry, 281(10), 6211–6218. https://doi.org/10.1074/jbc.M508582200
Xiong, Y., Collins, Q. F., An, J., Lupo, E., Jr., Liu, H. Y., Liu, D., Robidoux, J., Liu, Z., & Cao, W. (2007). p38 mitogen-activated protein kinase plays an inhibitory role in hepatic lipogenesis. Journal of Biological Chemistry, 282(7), 4975–4982. https://doi.org/10.1074/jbc.M606742200
London, E., & Stratakis, C. A. (2022). The regulation of PKA signaling in obesity and in the maintenance of metabolic health. Pharmacology & Therapeutics, 237, 108113. https://doi.org/10.1016/j.pharmthera.2022.108113
Yan, F., Wang, Q., Lu, M., Chen, W., Song, Y., Jing, F., Guan, Y., Wang, L., Lin, Y., Bo, T., Zhang, J., Wang, T., Xin, W., Yu, C., Guan, Q., Zhou, X., Gao, L., Xu, C., & Zhao, J. (2014). Thyrotropin increases hepatic triglyceride content through upregulation of SREBP-1c activity. Journal of Hepatology, 61(6), 1358–1364. https://doi.org/10.1016/j.jhep.2014.06.037
Rochira, A., Damiano, F., Marsigliante, S., Gnoni, G. V., & Siculella, L. (2013). 3,5-Diiodo-l-thyronine induces SREBP-1 proteolytic cleavage block and apoptosis in human hepatoma (Hepg2) cells. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids, 1831(12), 1679–1689. https://doi.org/10.1016/j.bbalip.2013.08.003
Gnoni, G. V., Rochira, A., Leone, A., Damiano, F., Marsigliante, S., & Siculella, L. (2012). 3,5,3′triiodo-L-thyronine induces SREBP-1 expression by non-genomic actions in human HEP G2 cells. Journal of Cellular Physiology, 227(6), 2388–2397. https://doi.org/10.1002/jcp.22974
Mueller, K. M., Kornfeld, J. W., Friedbichler, K., Blaas, L., Egger, G., Esterbauer, H., Hasselblatt, P., Schlederer, M., Haindl, S., Wagner, K. U., Engblom, D., Haemmerle, G., Kratky, D., Sexl, V., Kenner, L., Kozlov, A. V., Terracciano, L., Zechner, R., Schuetz, G., et al. (2011). Impairment of hepatic growth hormone and glucocorticoid receptor signaling causes steatosis and hepatocellular carcinoma in mice. Hepatology, 54(4), 1398–1409. https://doi.org/10.1002/hep.24509
Seidu, T., McWhorter, P., Myer, J., Alamgir, R., Eregha, N., Bogle, D., Lofton, T., Ecelbarger, C., & Andrisse, S. (2021). DHT causes liver steatosis via transcriptional regulation of SCAP in normal weight female mice. The Journal of Endocrinology, 250(2), 49–65. https://doi.org/10.1530/joe-21-0040
Min, X., Wen, J., Zhao, L., Wang, K., Li, Q., Huang, G., Liu, J., & Zhao, X. (2018). Role of hepatoma-derived growth factor in promoting de novo lipogenesis and tumorigenesis in hepatocellular carcinoma. Molecular Oncology, 12(9), 1480–1497. https://doi.org/10.1002/1878-0261.12357
Li, Y., Wong, K., Giles, A., Jiang, J., Lee, J. W., Adams, A. C., Kharitonenkov, A., Yang, Q., Gao, B., Guarente, L., & Zang, M. (2014). Hepatic SIRT1 attenuates hepatic steatosis and controls energy balance in mice by inducing fibroblast growth factor 21. Gastroenterology, 146(2), 539–549.e537. https://doi.org/10.1053/j.gastro.2013.10.059
Zhang, Y., Lei, T., Huang, J. F., Wang, S. B., Zhou, L. L., Yang, Z. Q., & Chen, X. D. (2011). The link between fibroblast growth factor 21 and sterol regulatory element binding protein 1c during lipogenesis in hepatocytes. Molecular and cellular Endocrinology, 342(1-2), 41–47. https://doi.org/10.1016/j.mce.2011.05.003
Ruscica, M., Ricci, C., Macchi, C., Magni, P., Cristofani, R., Liu, J., Corsini, A., & Ferri, N. (2016). Suppressor of cytokine signaling-3 (SOCS-3) induces proprotein convertase subtilisin kexin type 9 (PCSK9) expression in hepatic HepG2 cell line. Journal of Biological Chemistry, 291(7), 3508–3519. https://doi.org/10.1074/jbc.M115.664706
Ma, H. Y., Yamamoto, G., Xu, J., Liu, X., Karin, D., Kim, J. Y., Alexandrov, L. B., Koyama, Y., Nishio, T., Benner, C., Heinz, S., Rosenthal, S. B., Liang, S., Sun, M., Karin, G., Zhao, P., Brodt, P., McKillop, I. H., Quehenberger, O., Dennis, E., et al. (2020). IL-17 signaling in steatotic hepatocytes and macrophages promotes hepatocellular carcinoma in alcohol-related liver disease. Journal of Hepatology, 72(5), 946-959. http://doi.org/https://doi.org/10.1016/j.jhep.2019.12.016
Lin, Y. X., Wu, X. B., Zheng, C. W., Zhang, Q. L., Zhang, G. Q., Chen, K., Zhan, Q., & An, F. M. (2021). Mechanistic investigation on the regulation of FABP1 by the IL-6/miR-603 signaling in the pathogenesis of hepatocellular carcinoma. BioMed Research International, 2021, 8579658. https://doi.org/10.1155/2021/8579658
Rao, S., Yang, X., Ohshiro, K., Zaidi, S., Wang, Z., Shetty, K., Xiang, X., Hassan, M. I., Mohammad, T., Latham, P. S., Nguyen, B. N., Wong, L., Yu, H., Al-Abed, Y., Mishra, B., Vacca, M., Guenigault, G., Allison, M. E. D., Vidal-Puig, A., et al. (2021). Beta2-spectrin (SPTBN1) as a therapeutic target for diet-induced liver disease and preventing cancer development. Science Translational Medicine, 13(624), eabk2267. https://doi.org/10.1126/scitranslmed.abk2267
Xiang, X., Ohshiro, K., Zaidi, S., Yang, X., Bhowmick, K., Vegesna, A. K., Bernstein, D., Crawford, J. M., Mishra, B., Latham, P. S., Gough, N. R., Rao, S., & Mishra, L. (2022). Impaired reciprocal regulation between SIRT6 and TGF-β signaling in fatty liver. FASEB Journal, 36(6), e22335. https://doi.org/10.1096/fj.202101518R
Watanabe, M., Houten, S. M., Wang, L., Moschetta, A., Mangelsdorf, D. J., Heyman, R. A., Moore, D. D., & Auwerx, J. (2004). Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. The Journal of Clinical Investigation, 113(10), 1408–1418. https://doi.org/10.1172/JCI21025
Wang, K., Zhang, Z., Tsai, H. I., Liu, Y., Gao, J., Wang, M., Song, L., Cao, X., Xu, Z., Chen, H., Gong, A., Wang, D., Cheng, F., & Zhu, H. (2021). Branched-chain amino acid aminotransferase 2 regulates ferroptotic cell death in cancer cells. Cell Death & Differentiation, 28(4), 1222–1236. https://doi.org/10.1038/s41418-020-00644-4
Takeuchi, Y., Yahagi, N., Izumida, Y., Nishi, M., Kubota, M., Teraoka, Y., Yamamoto, T., Matsuzaka, T., Nakagawa, Y., Sekiya, M., Iizuka, Y., Ohashi, K., Osuga, J., Gotoda, T., Ishibashi, S., Itaka, K., Kataoka, K., Nagai, R., Yamada, N., et al. (2010). Polyunsaturated fatty acids selectively suppress sterol regulatory element-binding protein-1 through proteolytic processing and autoloop regulatory circuit. Journal of Biological Chemistry, 285(15), 11681–11691. https://doi.org/10.1074/jbc.M109.096107
Yoshikawa, T., Shimano, H., Yahagi, N., Ide, T., Amemiya-Kudo, M., Matsuzaka, T., Nakakuki, M., Tomita, S., Okazaki, H., Tamura, Y., Iizuka, Y., Ohashi, K., Takahashi, A., Sone, H., Osuga Ji, J., Gotoda, T., Ishibashi, S., & Yamada, N. (2002). Polyunsaturated fatty acids suppress sterol regulatory element-binding protein 1c promoter activity by inhibition of liver X receptor (LXR) binding to LXR response elements. The Journal of biological chemistry, 277(3), 1705–1711. https://doi.org/10.1074/jbc.M105711200
Xu, J., Teran-Garcia, M., Park, J. H., Nakamura, M. T., & Clarke, S. D. (2001). Polyunsaturated fatty acids suppress hepatic sterol regulatory element-binding protein-1 expression by accelerating transcript decay. The Journal of biological chemistry, 276(13), 9800–9807. https://doi.org/10.1074/jbc.M008973200
Hoxhaj, G., & Manning, B. D. (2020). The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nature Reviews. Cancer, 20(2), 74–88. https://doi.org/10.1038/s41568-019-0216-7
Jones, S. A., & Jenkins, B. J. (2018). Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nature Reviews. Immunology, 18(12), 773–789. https://doi.org/10.1038/s41577-018-0066-7
Derynck, R., Turley, S. J., & Akhurst, R. J. (2021). TGFβ biology in cancer progression and immunotherapy. Nature Reviews. Clinical Oncology, 18(1), 9–34. https://doi.org/10.1038/s41571-020-0403-1
Peng, W. T., Sun, W. Y., Li, X. R., Sun, J. C., Du, J. J., & Wei, W. (2018). Emerging roles of G protein-coupled receptors in hepatocellular carcinoma. Intertional Journal of Molecular Sciences, 19(5). https://doi.org/10.3390/ijms19051366
Wacker, D., Stevens, R. C., & Roth, B. L. (2017). How ligands illuminate GPCR molecular pharmacology. Cell, 170(3), 414–427. https://doi.org/10.1016/j.cell.2017.07.009
Trefts, E., & Shaw, R. J. (2021). AMPK: Restoring metabolic homeostasis over space and time. Molecular Cell, 81(18), 3677–3690. https://doi.org/10.1016/j.molcel.2021.08.015
Georgiadi, A., & Kersten, S. (2012). Mechanisms of gene regulation by fatty acids. Advances in Nutrition, 3(2), 127–134. https://doi.org/10.3945/an.111.001602
Sun, L., Cai, J., & Gonzalez, F. J. (2021). The role of farnesoid X receptor in metabolic diseases, and gastrointestinal and liver cancer. Nature Reviews. Gastroenterology & Hepatology, 18(5), 335–347. https://doi.org/10.1038/s41575-020-00404-2
Bovenga, F., Sabbà, C., & Moschetta, A. (2015). Uncoupling nuclear receptor LXR and cholesterol metabolism in cancer. Cell Metabolism, 21(4), 517–526. https://doi.org/10.1016/j.cmet.2015.03.002
Yue, S., Li, G., He, S., & Li, T. (2022). The central role of mTORC1 in amino acid sensing. Cancer Research, 82(17), 2964–2974. https://doi.org/10.1158/0008-5472.CAN-21-4403
Sundqvist, A., Bengoechea-Alonso, M. T., Ye, X., Lukiyanchuk, V., Jin, J., Harper, J. W., & Ericsson, J. (2005). Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7). Cell Metabolism, 1(6), 379–391. https://doi.org/10.1016/j.cmet.2005.04.010
Koeberle, S. C., Kipp, A. P., Stuppner, H., & Koeberle, A. (2023). Ferroptosis-modulating small molecules for targeting drug-resistant cancer: Challenges and opportunities in manipulating redox signaling. Medicinal Research Reviews. https://doi.org/10.1002/med.21933
Galbraith, L., Leung, H. Y., & Ahmad, I. (2018). Lipid pathway deregulation in advanced prostate cancer. Pharmacological Research, 131, 177–184. https://doi.org/10.1016/j.phrs.2018.02.022
An, H. J., Kim, J. Y., Gwon, M. G., Gu, H., Kim, H. J., Leem, J., Youn, S. W., & Park, K. K. (2020). Beneficial effects of SREBP decoy oligodeoxynucleotide in an animal model of hyperlipidemia. Intertional Journal of Molecular Sciences, 21(2). https://doi.org/10.3390/ijms21020552
Tang, J. J., Li, J. G., Qi, W., Qiu, W. W., Li, P. S., Li, B. L., & Song, B. L. (2011). Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metabolism, 13(1), 44–56. https://doi.org/10.1016/j.cmet.2010.12.004
Kamisuki, S., Mao, Q., Abu-Elheiga, L., Gu, Z., Kugimiya, A., Kwon, Y., Shinohara, T., Kawazoe, Y., Sato, S.-I., Asakura, K., Choo, H.-Y. P., Sakai, J., Wakil, S. J., & Uesugi, M. (2009). A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chemistry & Biology, 16(8), 882–892. https://doi.org/10.1016/j.chembiol.2009.07.007
Zheng, Z. G., Zhu, S. T., Cheng, H. M., Zhang, X., Cheng, G., Thu, P. M., Wang, S. P., Li, H. J., Ding, M., Qiang, L., Chen, X. W., Zhong, Q., Li, P., & Xu, X. (2021). Discovery of a potent SCAP degrader that ameliorates HFD-induced obesity, hyperlipidemia and insulin resistance via an autophagy-independent lysosomal pathway. Autophagy, 17(7), 1592–1613. https://doi.org/10.1080/15548627.2020.1757955
Jensen, K. K., Tadin-Strapps, M., Wang, S. P., Hubert, J., Kan, Y., Ma, Y., McLaren, D. G., Previs, S. F., Herath, K. B., Mahsut, A., Liaw, A., Wang, S., Stout, S. J., Keohan, C., Forrest, G., Coelho, D., Yendluri, S., Williams, S., Koser, M., et al. (2016). Dose-dependent effects of siRNA-mediated inhibition of SCAP on PCSK9, LDLR, and plasma lipids in mouse and rhesus monkey. Journal of Lipid Research, 57(12), 2150–2162. https://doi.org/10.1194/jlr.M071498
Murphy, B. A., Tadin-Strapps, M., Jensen, K., Mogg, R., Liaw, A., Herath, K., Bhat, G., McLaren, D. G., Previs, S. F., & Pinto, S. (2017). siRNA-mediated inhibition of SREBP cleavage-activating protein reduces dyslipidemia in spontaneously dysmetabolic rhesus monkeys. Metabolism: Clinical and Experimental, 71, 202–212. https://doi.org/10.1016/j.metabol.2017.02.015
Miyata, S., Inoue, J., Shimizu, M., & Sato, R. (2015). Xanthohumol improves diet-induced obesity and fatty liver by suppressing sterol regulatory element-binding protein (SREBP) activation. Journal of Biological Chemistry, 290(33), 20565–20579. https://doi.org/10.1074/jbc.M115.656975
Miyata, S., Kodaka, M., Kikuchi, A., Matsunaga, Y., Shoji, K., Kuan, Y. C., Iwase, M., Takeda, K., Katsuta, R., Ishigami, K., Matsumoto, Y., Suzuki, T., Yamamoto, Y., Sato, R., & Inoue, J. (2022). Sulforaphane suppresses the activity of sterol regulatory element-binding proteins (SREBPs) by promoting SREBP precursor degradation. Scientific Reports, 12(1), 8715. https://doi.org/10.1038/s41598-022-12347-6
Tian, S., Li, B., Lei, P., Yang, X., Zhang, X., Bao, Y., & Shan, Y. (2018). Sulforaphane improves abnormal lipid metabolism via both ERS-dependent XBP1/ACC &SCD1 and ERS-independent SREBP/FAS pathways. Molecular Nutrition & Food Research, 62(6), e1700737. https://doi.org/10.1002/mnfr.201700737
Zhang, C., Sheng, L., Yuan, M., Hu, J., Meng, Y., Wu, Y., Chen, L., Yu, H., Li, S., Zheng, G., & Qiu, Z. (2020). Orlistat delays hepatocarcinogenesis in mice with hepatic co-activation of AKT and c-Met. Toxicology and Applied Pharmacology, 392, 114918. https://doi.org/10.1016/j.taap.2020.114918
Liu, T., Yu, J., Ge, C., Zhao, F., Chen, J., Miao, C., Jin, W., Zhou, Q., Geng, Q., Lin, H., Tian, H., Chen, T., Xie, H., Cui, Y., Yao, M., Xiao, X., Li, J., & Li, H. (2022). Sperm associated antigen 4 promotes SREBP1-mediated de novo lipogenesis via interaction with lamin A/C and contributes to tumor progression in hepatocellular carcinoma. Cancer Letters, 536, 215642. https://doi.org/10.1016/j.canlet.2022.215642
Pai, S. A., Munshi, R. P., Panchal, F. H., Gaur, I. S., Mestry, S. N., Gursahani, M. S., & Juvekar, A. R. (2019). Plumbagin reduces obesity and nonalcoholic fatty liver disease induced by fructose in rats through regulation of lipid metabolism, inflammation and oxidative stress. Biomedicine & Pharmacotherapy, 111, 686–694. https://doi.org/10.1016/j.biopha.2018.12.139
Wu, J.-B., Kuo, Y.-H., Lin, C.-H., Ho, H.-Y., & Shih, C.-C. (2014). Tormentic acid, a major component of suspension cells of Eriobotrya japonica, suppresses high-fat diet-induced diabetes and hyperlipidemia by glucose transporter 4 and AMP-activated protein kinase phosphorylation. Journal of Agricultural and Food Chemistry, 62(44), 10717–10726. https://doi.org/10.1021/jf503334d
Yang, Q., Mao, Y., Wang, J., Yu, H., Zhang, X., Pei, X., Duan, Z., Xiao, C., & Ma, M. (2022). Gestational bisphenol A exposure impairs hepatic lipid metabolism by altering mTOR/CRTC2/SREBP1 in male rat offspring. Human & Experimental Toxicology, 41, 9603271221129852. https://doi.org/10.1177/09603271221129852
Zheng, L., Yang, W., Wu, F., Wang, C., Yu, L., Tang, L., Qiu, B., Li, Y., Guo, L., Wu, M., Feng, G., Zou, D., & Wang, H. (2013). Prognostic significance of AMPK activation and therapeutic effects of metformin in hepatocellular carcinoma. Clinical Cancer Research, 19(19), 5372–5380. https://doi.org/10.1158/1078-0432.CCR-13-0203
Chen, H. P., Shieh, J. J., Chang, C. C., Chen, T. T., Lin, J. T., Wu, M. S., Lin, J. H., & Wu, C. Y. (2013). Metformin decreases hepatocellular carcinoma risk in a dose-dependent manner: Population-based and in vitro studies. Gut, 62(4), 606–615. https://doi.org/10.1136/gutjnl-2011-301708
Donadon, V., Balbi, M., Mas, M. D., Casarin, P., & Zanette, G. (2010). Metformin and reduced risk of hepatocellular carcinoma in diabetic patients with chronic liver disease. Liver Internatonal, 30(5), 750–758. https://doi.org/10.1111/j.1478-3231.2010.02223.x
You, M., Matsumoto, M., Pacold, C. M., Cho, W. K., & Crabb, D. W. (2004). The role of AMP-activated protein kinase in the action of ethanol in the liver. Gastroenterology, 127(6), 1798–1808. https://doi.org/10.1053/j.gastro.2004.09.049
Zang, M., Xu, S., Maitland-Toolan, K. A., Zuccollo, A., Hou, X., Jiang, B., Wierzbicki, M., Verbeuren, T. J., & Cohen, R. A. (2006). Polyphenols stimulate AMP-activated protein kinase, lower lipids, and inhibit accelerated atherosclerosis in diabetic LDL receptor–deficient mice. Diabetes, 55(8), 2180–2191. https://doi.org/10.2337/db05-1188
Li, Y., Xu, S., Mihaylova, M. M., Zheng, B., Hou, X., Jiang, B., Park, O., Luo, Z., Lefai, E., Shyy, J. Y., Gao, B., Wierzbicki, M., Verbeuren, T. J., Shaw, R. J., Cohen, R. A., & Zang, M. (2011). AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metabolism, 13(4), 376–388. https://doi.org/10.1016/j.cmet.2011.03.009
Han, Y., Hu, Z., Cui, A., Liu, Z., Ma, F., Xue, Y., Liu, Y., Zhang, F., Zhao, Z., Yu, Y., Gao, J., Wei, C., Li, J., Fang, J., Li, J., Fan, J. G., Song, B. L., & Li, Y. (2019). Post-translational regulation of lipogenesis via AMPK-dependent phosphorylation of insulin-induced gene. Nature Communications, 10(1), 623. https://doi.org/10.1038/s41467-019-08585-4
Cheng, L., Deepak, R., Wang, G., Meng, Z., Tao, L., Xie, M., Chi, W., Zhang, Y., Yang, M., Liao, Y., Chen, R., Liang, Y., Zhang, J., Huang, Y., Wang, W., Guo, Z., Wang, Y., Lin, J. D., Fan, H., & Chen, L. (2023). Hepatic mitochondrial NAD + transporter SLC25A47 activates AMPKα mediating lipid metabolism and tumorigenesis. Hepatology. https://doi.org/10.1097/HEP.0000000000000314
Cao, K., Xu, J., Zou, X., Li, Y., Chen, C., Zheng, A., Li, H., Li, H., Szeto, I. M.-Y., Shi, Y., Long, J., Liu, J., & Feng, Z. (2014). Hydroxytyrosol prevents diet-induced metabolic syndrome and attenuates mitochondrial abnormalities in obese mice. Free Radical Biology and Medicine, 67, 396–407. https://doi.org/10.1016/j.freeradbiomed.2013.11.029
Souza-Mello, V., Gregório, B. M., Cardoso-de-Lemos, F. S., de Carvalho, L., Aguila, M. B., & Mandarim-de-Lacerda, C. A. (2010). Comparative effects of telmisartan, sitagliptin and metformin alone or in combination on obesity, insulin resistance, and liver and pancreas remodelling in C57BL/6 mice fed on a very high-fat diet. Clinical Science, 119(6), 239–250. https://doi.org/10.1042/CS20100061
Jung, E. J., Kwon, S. W., Jung, B. H., Oh, S. H., & Lee, B. H. (2011). Role of the AMPK/SREBP-1 pathway in the development of orotic acid-induced fatty liver. Journal of Lipid Research, 52(9), 1617–1625. https://doi.org/10.1194/jlr.M015263
Ma, M., Duan, R., Shen, L., Liu, M., Ji, Y., Zhou, H., Li, C., Liang, T., Li, X., & Guo, L. (2020). The lncRNA Gm15622 stimulates SREBP-1c expression and hepatic lipid accumulation by sponging the miR-742-3p in mice. Journal of Lipid Research, 61(7), 1052–1064. https://doi.org/10.1194/jlr.RA120000664
Cai, X., Liu, Z., Dong, X., Wang, Y., Zhu, L., Li, M., & Xu, Y. (2021). Hypoglycemic and lipid lowering effects of theaflavins in high-fat diet-induced obese mice. Food & Function, 12(20), 9922–9931. https://doi.org/10.1039/D1FO01966J
Bai, T., Yang, Y., Yao, Y. L., Sun, P., Lian, L. H., Wu, Y. L., & Nan, J. X. (2016). Betulin alleviated ethanol-induced alcoholic liver injury via SIRT1/AMPK signaling pathway. Pharmacological Research, 105, 1–12. https://doi.org/10.1016/j.phrs.2015.12.022
Liu, G., Kuang, S., Cao, R., Wang, J., Peng, Q., & Sun, C. (2019). Sorafenib kills liver cancer cells by disrupting SCD1-mediated synthesis of monounsaturated fatty acids via the ATP-AMPK-mTOR-SREBP1 signaling pathway. The FASEB Journal, 33(9), 10089–10103. https://doi.org/10.1096/fj.201802619RR
Choi, Y. J., Suh, H. R., Yoon, Y., Lee, K. J., Kim, D. G., Kim, S., & Lee, B. H. (2014). Protective effect of resveratrol derivatives on high-fat diet induced fatty liver by activating AMP-activated protein kinase. Archives of Pharmacal Research, 37(9), 1169–1176. https://doi.org/10.1007/s12272-014-0347-z
Alberdi, G., Rodríguez, V. M., Macarulla, M. T., Miranda, J., Churruca, I., & Portillo, M. P. (2013). Hepatic lipid metabolic pathways modified by resveratrol in rats fed an obesogenic diet. Nutrition, 29(3), 562–567. https://doi.org/10.1016/j.nut.2012.09.011
Ajmo, J. M., Liang, X., Rogers, C. Q., Pennock, B., & You, M. (2008). Resveratrol alleviates alcoholic fatty liver in mice. American Journal of Physiology. Gastrointestinal and Liver Physiology, 295(4), G833–G842. https://doi.org/10.1152/ajpgi.90358.2008
Lin, H. C., Chen, Y. F., Hsu, W. H., Yang, C. W., Kao, C. H., & Tsai, T. F. (2012). Resveratrol helps recovery from fatty liver and protects against hepatocellular carcinoma induced by hepatitis B virus X protein in a mouse model. Cancer Prevention Research, 5(7), 952–962. https://doi.org/10.1158/1940-6207.CAPR-12-0001
Zhou, J., Yoshitomi, H., Liu, T., Zhou, B., Sun, W., Qin, L., Guo, X., Huang, L., Wu, L., & Gao, M. (2014). Isoquercitrin activates the AMP-activated protein kinase (AMPK) signal pathway in rat H4IIE cells. BMC Complementary and Alternative Medicine, 14, 42. https://doi.org/10.1186/1472-6882-14-42
Manzoor, M., Muroi, M., Ogawa, N., Kobayashi, H., Nishimura, H., Chen, D., Fasina, O. B., Wang, J., Osada, H., Yoshida, M., Xiang, L., & Qi, J. (2022). Isoquercitrin from Apocynum venetum L. produces an anti-obesity effect on obese mice by targeting C-1-tetrahydrofolate synthase, carbonyl reductase, and glutathione S-transferase P and modification of the AMPK/SREBP-1c/FAS/CD36 signaling pathway in mice in vivo. Food & Function, 13(21), 10923–10936. https://doi.org/10.1039/D2FO02438A
Kim, S. H., Yun, C., Kwon, D., Lee, Y. H., Kwak, J. H., & Jung, Y. S. (2023). Effect of isoquercitrin on free fatty acid-induced lipid accumulation in HepG2 cells. Molecules, 28(3). https://doi.org/10.3390/molecules28031476
Kang, O. H., Kim, S. B., Seo, Y. S., Joung, D. K., Mun, S. H., Choi, J. G., Lee, Y. M., Kang, D. G., Lee, H. S., & Kwon, D. Y. (2013). Curcumin decreases oleic acid-induced lipid accumulation via AMPK phosphorylation in hepatocarcinoma cells. European Review for Medical and Pharmacological Sciences, 17(19), 2578–2586.
Liu, Z., Cui, C., Xu, P., Dang, R., Cai, H., Liao, D., Yang, M., Feng, Q., Yan, X., & Jiang, P. (2017). Curcumin activates AMPK pathway and regulates lipid metabolism in rats following prolonged clozapine exposure. Frontiers in Neuroscience, 11, 558. https://doi.org/10.3389/fnins.2017.00558
Zhou, B., Zhou, D. L., Wei, X. H., Zhong, R. Y., Xu, J., & Sun, L. (2017). Astragaloside IV attenuates free fatty acid-induced ER stress and lipid accumulation in hepatocytes via AMPK activation. Acta Pharmacologica Sinica, 38(7), 998–1008. https://doi.org/10.1038/aps.2016.175
Wang, C., Li, Y., Hao, M., & Li, W. (2018). Astragaloside IV inhibits triglyceride accumulation in insulin-resistant HepG2 cells via AMPK-induced SREBP-1c phosphorylation. Frontiers in Pharmacology, 9, 345. https://doi.org/10.3389/fphar.2018.00345
Wang, S. W., Lan, T., Chen, H. F., Sheng, H., Xu, C. Y., Xu, L. F., Zheng, F., & Zhang, F. (2022). Limonin, an AMPK activator, inhibits hepatic lipid accumulation in high fat diet fed mice. Frontiers in Pharmacology, 13, 833705. https://doi.org/10.3389/fphar.2022.833705
Xu, H., Lyu, X., Guo, X., Yang, H., Duan, L., Zhu, H., Pan, H., Gong, F., & Wang, L. (2022). Distinct AMPK-mediated FAS/HSL pathway is implicated in the alleviating effect of nuciferine on obesity and hepatic steatosis in HFD-fed mice. Nutrients, 14(9). https://doi.org/10.3390/nu14091898
Guo, L., Kang, J. S., Park, Y. H., Je, B. I., Lee, Y. J., Kang, N. J., Park, S. Y., Hwang, D. Y., & Choi, Y. W. (2020). S-Petasin inhibits lipid accumulation in oleic acid-induced HepG2 cells through activation of the AMPK signaling pathway. Food & Function, 11(6), 5664–5673. https://doi.org/10.1039/D0FO00594K
Hwang, Y. P., Kim, H. G., Choi, J. H., Do, M. T., Chung, Y. C., Jeong, T. C., & Jeong, H. G. (2013). S-allyl cysteine attenuates free fatty acid-induced lipogenesis in human HepG2 cells through activation of the AMP-activated protein kinase-dependent pathway. The Journal of Nutritional Biochemistry, 24(8), 1469–1478. https://doi.org/10.1016/j.jnutbio.2012.12.006
Takemura, S., Minamiyama, Y., Kodai, S., Shinkawa, H., Tsukioka, T., Okada, S., Azuma, H., & Kubo, S. (2013). S-Allyl cysteine improves nonalcoholic fatty liver disease in type 2 diabetes Otsuka Long-Evans Tokushima Fatty rats via regulation of hepatic lipogenesis and glucose metabolism. Journal of Clinical Biochemistry and Nutrition, 53(2), 94–101. https://doi.org/10.3164/jcbn.13-1
Quan, H. Y., Kim, D. Y., Kim, S. J., Jo, H. K., Kim, G. W., & Chung, S. H. (2013). Betulinic acid alleviates non-alcoholic fatty liver by inhibiting SREBP1 activity via the AMPK-mTOR-SREBP signaling pathway. Biochemical Pharmacology, 85(9), 1330–1340. https://doi.org/10.1016/j.bcp.2013.02.007
Wang, D., Wei, Y., Wang, T., Wan, X., Yang, C. S., Reiter, R. J., & Zhang, J. (2015). Melatonin attenuates (-)-epigallocatehin-3-gallate-triggered hepatotoxicity without compromising its downregulation of hepatic gluconeogenic and lipogenic genes in mice. Journal of Pineal Research, 59(4), 497–507. https://doi.org/10.1111/jpi.12281
Lu, J., Fang, B., Huang, Y., Tao, S., Sun, B., Guan, S., & Jin, Y. (2018). Epigallocatechin-3-gallate protects against 1,3-dichloro-2-propanol-induced lipid accumulation in C57BL/6J mice. Life Sciences, 209, 324–331. https://doi.org/10.1016/j.lfs.2018.08.007
Ding, H., Li, Y., Li, W., Tao, H., Liu, L., Zhang, C., Kong, T., Feng, S., Li, J., Wang, X., & Wu, J. (2021). Epigallocatechin-3-gallate activates the AMP-activated protein kinase signaling pathway to reduce lipid accumulation in canine hepatocytes. Journal of Cellular Physiologz, 236(1), 405–416. https://doi.org/10.1002/jcp.29869
Lee, M. S., Kim, D., Jo, K., & Hwang, J. K. (2010). Nordihydroguaiaretic acid protects against high-fat diet-induced fatty liver by activating AMP-activated protein kinase in obese mice. Biochemical and Biophysical Research Communications, 401(1), 92–97. https://doi.org/10.1016/j.bbrc.2010.09.016
Zhang, H., Li, Y., Hu, J., Shen, W. J., Singh, M., Hou, X., Bittner, A., Bittner, S., Cortez, Y., Tabassum, J., Kraemer, F. B., & Azhar, S. (2015). Effect of creosote bush-derived NDGA on expression of genes involved in lipid metabolism in liver of high-fructose fed rats: relevance to NDGA amelioration of hypertriglyceridemia and hepatic steatosis. PLoS One, 10(9), e0138203. https://doi.org/10.1371/journal.pone.0138203
Yang, J. W., Kim, H. S., Im, J. H., Kim, J. W., Jun, D. W., Lim, S. C., Lee, K., Choi, J. M., Kim, S. K., & Kang, K. W. (2016). GPR119: A promising target for nonalcoholic fatty liver disease. The FASEB Journal, 30(1), 324–335. https://doi.org/10.1096/fj.15-273771
Long, Q., Chen, H., Yang, W., Yang, L., & Zhang, L. (2021). Delphinidin-3-sambubioside from Hibiscus sabdariffa. L attenuates hyperlipidemia in high fat diet-induced obese rats and oleic acid-induced steatosis in HepG2 cells. Bioengineered, 12(1), 3837–3849. https://doi.org/10.1080/21655979.2021.1950259
Lin, Y. N., Wang, C. C. N., Chang, H. Y., Chu, F. Y., Hsu, Y. A., Cheng, W. K., Ma, W. C., Chen, C. J., Wan, L., & Lim, Y. P. (2018). Ursolic acid, a novel liver X receptor α (LXRα) antagonist inhibiting ligand-induced nonalcoholic fatty liver and drug-induced lipogenesis. Journal of Agricultural and Food Chemistry, 66(44), 11647–11662. https://doi.org/10.1021/acs.jafc.8b04116
Liu, C., Shen, Y. J., Tu, Q. B., Zhao, Y. R., Guo, H., Wang, J., Zhang, L., Shi, H. W., & Sun, Y. (2018). Pedunculoside, a novel triterpene saponin extracted from Ilex rotunda, ameliorates high-fat diet induced hyperlipidemia in rats. Biomedicine & Pharmacotherapy, 101, 608–616. https://doi.org/10.1016/j.biopha.2018.02.131
Kim, J. H., Lee, J. M., Kim, J. H., & Kim, K. R. (2018). Fluvastatin activates sirtuin 6 to regulate sterol regulatory element-binding proteins and AMP-activated protein kinase in HepG2 cells. Biochemical and Biophysical Research Communications, 503(3), 1415–1421. https://doi.org/10.1016/j.bbrc.2018.07.057
Kim, H. Y., Hong, M. H., Kim, K. W., Yoon, J. J., Lee, J. E., Kang, D. G., & Lee, H. S. (2020). Improvement of hypertriglyceridemia by roasted nelumbinis folium in high fat/high cholesterol diet rat model. Nutrients, 12(12). https://doi.org/10.3390/nu12123859
Liu, M., Zhang, G., Wu, S., Song, M., Wang, J., Cai, W., Mi, S., & Liu, C. (2020). Schaftoside alleviates HFD-induced hepatic lipid accumulation in mice via upregulating farnesoid X receptor. Journal of Ethnopharmacology, 255, 112776. https://doi.org/10.1016/j.jep.2020.112776
Zhang, Q., Yuan, H., Zhang, C., Guan, Y., Wu, Y., Ling, F., Niu, Y., & Li, Y. (2018). Epigallocatechin gallate improves insulin resistance in HepG2 cells through alleviating inflammation and lipotoxicity. Diabetes Research and Clinical Practice, 142, 363–373. https://doi.org/10.1016/j.diabres.2018.06.017
Roth, A., Looser, R., Kaufmann, M., Blättler, S. M., Rencurel, F., Huang, W., Moore, D. D., & Meyer, U. A. (2008). Regulatory cross-talk between drug metabolism and lipid homeostasis: constitutive androstane receptor and pregnane X receptor increase Insig-1 expression. Molecular pharmacology, 73(4), 1282–1289. https://doi.org/10.1124/mol.107.041012
Li, X., Li, S., Chen, M., Wang, J., Xie, B., & Sun, Z. (2018). (-)-Epigallocatechin-3-gallate (EGCG) inhibits starch digestion and improves glucose homeostasis through direct or indirect activation of PXR/CAR-mediated phase II metabolism in diabetic mice. Food & Function, 9(9), 4651–4663. https://doi.org/10.1039/C8FO01293H
Trasino, S. E., Tang, X. H., Jessurun, J., & Gudas, L. J. (2016). Retinoic acid receptor β2 agonists restore glycaemic control in diabetes and reduce steatosis. Diabetes Obesity and Metabolism, 18(2), 142–151. https://doi.org/10.1111/dom.12590
Wu, D., Li, J., Fan, Z., Wang, L., & Zheng, X. (2022). Resveratrol ameliorates oxidative stress, inflammatory response and lipid metabolism in common carp (Cyprinus carpio) fed with high-fat diet. Frontiers in Immunology, 13, 965954. https://doi.org/10.3389/fimmu.2022.965954
Damiano, F., Gnoni, G. V., & Siculella, L. (2012). Citrate carrier promoter is target of peroxisome proliferator-activated receptor alpha and gamma in hepatocytes and adipocytes. The International Journal of Biochemistry & Cell Biology, 44(4), 659–668. https://doi.org/10.1016/j.biocel.2012.01.003
Li, M., Meng, X., Xu, J., Huang, X., Li, H., Li, G., Wang, S., Man, Y., Tang, W., & Li, J. (2016). GPR40 agonist ameliorates liver X receptor-induced lipid accumulation in liver by activating AMPK pathway. Scientific Reports, 6, 25237. https://doi.org/10.1038/srep25237
On, S., Kim, H. Y., Kim, H. S., Park, J., & Kang, K. W. (2019). Involvement of G-protein-coupled receptor 40 in the inhibitory effects of docosahexaenoic acid on SREBP1-mediated lipogenic enzyme expression in primary hepatocytes. International Journal of Molecular Sciences, 20(11). https://doi.org/10.3390/ijms20112625
Zhao, X., Zhao, L., Yang, H., Li, J., Min, X., Yang, F., Liu, J., & Huang, G. (2018). Pyruvate kinase M2 interacts with nuclear sterol regulatory element-binding protein 1a and thereby activates lipogenesis and cell proliferation in hepatocellular carcinoma. Journal of Biological Chemistry, 293(17), 6623–6634. https://doi.org/10.1074/jbc.RA117.000100
Ponugoti, B., Kim, D. H., Xiao, Z., Smith, Z., Miao, J., Zang, M., Wu, S. Y., Chiang, C. M., Veenstra, T. D., & Kemper, J. K. (2010). SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. Journal of Biological Chemistry, 285(44), 33959–33970. https://doi.org/10.1074/jbc.M110.122978
Wu, W. Y., Ding, X. Q., Gu, T. T., Guo, W. J., Jiao, R. Q., Song, L., Sun, Y., Pan, Y., & Kong, L. D. (2020). Pterostilbene improves hepatic lipid accumulation via the MiR-34a/Sirt1/SREBP-1 pathway in fructose-fed rats. Journal of Agricultural and Food Chemistry, 68(5), 1436–1446. https://doi.org/10.1021/acs.jafc.2c00641
Tsai, H. Y., Shih, Y. Y., Yeh, Y. T., Huang, C. H., Liao, C. A., Hu, C. Y., Nagabhushanam, K., Ho, C. T., & Chen, Y. K. (2022). Pterostilbene and its derivative 3′-hydroxypterostilbene ameliorated nonalcoholic fatty liver disease through synergistic modulation of the gut microbiota and SIRT1/AMPK signaling pathway. Journal of Agricultural and Food Chemistry, 70(16), 4966–4980. https://doi.org/10.1021/acs.jafc.2c00641
Ju, U.-I., Jeong, D.-W., Seo, J., Park, J. B., Park, J.-W., Suh, K.-S., Kim, J. B., & Chun, Y.-S. (2020). Neddylation of sterol regulatory element-binding protein 1c is a potential therapeutic target for nonalcoholic fatty liver treatment. Cell Death & Disease, 11(4), 283. https://doi.org/10.1038/s41419-020-2472-6
Blanchet, M., Sureau, C., Guévin, C., Seidah, N. G., & Labonté, P. (2015). SKI-1/S1P inhibitor PF-429242 impairs the onset of HCV infection. Antiviral Research, 115, 94–104. https://doi.org/10.1016/j.antiviral.2014.12.017
Hwang, H. J., Jung, T. W., Kim, B. H., Hong, H. C., Seo, J. A., Kim, S. G., Kim, N. H., Choi, K. M., Choi, D. S., Baik, S. H., & Yoo, H. J. (2015). A dipeptidyl peptidase-IV inhibitor improves hepatic steatosis and insulin resistance by AMPK-dependent and JNK-dependent inhibition of LECT2 expression. Biochemical Pharmacology, 98(1), 157–166. https://doi.org/10.1016/j.bcp.2015.08.098
Wang, X., Hausding, M., Weng, S. Y., Kim, Y. O., Steven, S., Klein, T., Daiber, A., & Schuppan, D. (2018). Gliptins suppress inflammatory macrophage activation to mitigate inflammation, fibrosis, oxidative stress, and vascular dysfunction in models of nonalcoholic steatohepatitis and liver fibrosis. Antioxidants & Redox Signaling, 28(2), 87–109. https://doi.org/10.1089/ars.2016.6953
Syed, G. H., & Siddiqui, A. (2011). Effects of hypolipidemic agent nordihydroguaiaretic acid on lipid droplets and hepatitis C virus. Hepatology, 54(6), 1936–1946. https://doi.org/10.1002/hep.24619
Bai, X., Ali, A., Wang, N., Liu, Z., Lv, Z., Zhang, Z., Zhao, X., Hao, H., Zhang, Y., & Rahman, F. U. (2022). Inhibition of SREBP-mediated lipid biosynthesis and activation of multiple anticancer mechanisms by platinum complexes: Ascribe possibilities of new antitumor strategies. European Journal of Medicinal Chemistry, 227, 113920. https://doi.org/10.1016/j.ejmech.2021.113920
Yang, J. S., Tongson, J., Kim, K. H., & Park, Y. (2020). Piceatannol attenuates fat accumulation and oxidative stress in steatosis-induced HepG2 cells. Current Research in Food Science, 3, 92–99. https://doi.org/10.1016/j.crfs.2020.03.008
Tung, Y.-C., Lin, Y.-H., Chen, H.-J., Chou, S.-C., Cheng, A.-C., Kalyanam, N., Ho, C.-T., & Pan, M.-H. (2016). Piceatannol exerts anti-obesity effects in C57BL/6 mice through modulating adipogenic proteins and gut microbiota. Molecules, 21(11). https://doi.org/10.3390/molecules21111419
Shao, Y., Yao, Z., Zhou, J., Yu, M., Chen, S., Yuan, Y., Han, L., Jiang, L., & Liu, J. (2022). A novel small compound TOIDC suppresses lipogenesis via SREBP1-dependent signaling to curb MAFLD. Nutrition & Metabolism, 19(1), 80. https://doi.org/10.1186/s12986-022-00713-0
Wu, Y., Ma, K. L., Zhang, Y., Wen, Y., Wang, G. H., Hu, Z. B., Liu, L., Lu, J., Chen, P. P., Ruan, X. Z., & Liu, B. C. (2016). Lipid disorder and intrahepatic renin-angiotensin system activation synergistically contribute to non-alcoholic fatty liver disease. Liver Internatonal, 36(10), 1525–1534. https://doi.org/10.1111/liv.13131
Manning, B. D., & Toker, A. (2017). AKT/PKB signaling: Navigating the network. Cell, 169(3), 381–405. https://doi.org/10.1016/j.cell.2017.04.001
Porstmann, T., Santos, C. R., Griffiths, B., Cully, M., Wu, M., Leevers, S., Griffiths, J. R., Chung, Y. L., & Schulze, A. (2008). SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metabolism, 8(3), 224–236. https://doi.org/10.1016/j.cmet.2008.07.007
Li, J., Huang, Q., Long, X., Zhang, J., Huang, X., Aa, J., Yang, H., Chen, Z., & Xing, J. (2015). CD147 reprograms fatty acid metabolism in hepatocellular carcinoma cells through Akt/mTOR/SREBP1c and P38/PPARalpha pathways. Journal of Hepatology, 63(6), 1378–1389. https://doi.org/10.1016/j.jhep.2015.07.039
Mayer, A. E., Löffler, M. C., Loza Valdés, A. E., Schmitz, W., El-Merahbi, R., Viera, J. T., Erk, M., Zhang, T., Braun, U., Heikenwalder, M., Leitges, M., Schulze, A., & Sumara, G. (2019). The kinase PKD3 provides negative feedback on cholesterol and triglyceride synthesis by suppressing insulin signaling. Science Signaling, 12(593). https://doi.org/10.1126/scisignal.aav9150
Yecies, J. L., Zhang, H. H., Menon, S., Liu, S., Yecies, D., Lipovsky, A. I., Gorgun, C., Kwiatkowski, D. J., Hotamisligil, G. S., Lee, C. H., & Manning, B. D. (2011). Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metabolism, 14(1), 21–32. https://doi.org/10.1016/j.cmet.2011.06.002
Peterson, T. R., Sengupta, S. S., Harris, T. E., Carmack, A. E., Kang, S. A., Balderas, E., Guertin, D. A., Madden, K. L., Carpenter, A. E., Finck, B. N., & Sabatini, D. M. (2011). mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell, 146(3), 408–420. https://doi.org/10.1016/j.cell.2011.06.034
Han, J., & Wang, Y. (2018). mTORC1 signaling in hepatic lipid metabolism. Protein & Cell, 9(2), 145–151. https://doi.org/10.1007/s13238-017-0409-3
Gosis, B. S., Wada, S., Thorsheim, C., Li, K., Jung, S., Rhoades, J. H., Yang, Y., Brandimarto, J., Li, L., Uehara, K., Jang, C., Lanza, M., Sanford, N. B., Bornstein, M. R., Jeong, S., Titchenell, P. M., Biddinger, S. B., & Arany, Z. (2022). Inhibition of nonalcoholic fatty liver disease in mice by selective inhibition of mTORC1. Science, 376(6590), eabf8271. https://doi.org/10.1126/science.abf8271
Zhu, J., Wang, H., & Jiang, X. (2022). mTORC1 beyond anabolic metabolism: Regulation of cell death. Journal of Cell Biology, 221(12). https://doi.org/10.1083/jcb.202208103
Han, J., Li, E., Chen, L., Zhang, Y., Wei, F., Liu, J., Deng, H., & Wang, Y. (2015). The CREB coactivator CRTC2 controls hepatic lipid metabolism by regulating SREBP1. Nature, 524(7564), 243–246. https://doi.org/10.1038/nature14557
Memmott, R. M., & Dennis, P. A. (2009). Akt-dependent and -independent mechanisms of mTOR regulation in cancer. Cellular Signalling, 21(5), 656–664. https://doi.org/10.1016/j.cellsig.2009.01.004
Battaglioni, S., Benjamin, D., Wälchli, M., Maier, T., & Hall, M. N. (2022). mTOR substrate phosphorylation in growth control. Cell, 185(11), 1814–1836. https://doi.org/10.1016/j.cell.2022.04.013
Menon, S., Yecies, J. L., Zhang, H. H., Howell, J. J., Nicholatos, J., Harputlugil, E., Bronson, R. T., Kwiatkowski, D. J., & Manning, B. D. (2012). Chronic activation of mTOR complex 1 is sufficient to cause hepatocellular carcinoma in mice. Science Signaling, 5(217), ra24. https://doi.org/10.1126/scisignal.2002739
Yu, S., Meng, S., Xiang, M., & Ma, H. (2021). Phosphoenolpyruvate carboxykinase in cell metabolism: Roles and mechanisms beyond gluconeogenesis. Molecular Metabolism, 53, 101257. https://doi.org/10.1016/j.molmet.2021.101257
Xu, D., Wang, Z., Xia, Y., Shao, F., Xia, W., Wei, Y., Li, X., Qian, X., Lee, J. H., Du, L., Zheng, Y., Lv, G., Leu, J. S., Wang, H., Xing, D., Liang, T., Hung, M. C., & Lu, Z. (2020). The gluconeogenic enzyme PCK1 phosphorylates INSIG1/2 for lipogenesis. Nature, 580(7804), 530–535. https://doi.org/10.1038/s41586-020-2183-2
Welcker, M., & Clurman, B. E. (2008). FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nature Reviews Cancer, 8(2), 83–93. https://doi.org/10.1038/nrc2290
Li, S., Oh, Y. T., Yue, P., Khuri, F. R., & Sun, S. Y. (2016). Inhibition of mTOR complex 2 induces GSK3/FBXW7-dependent degradation of sterol regulatory element-binding protein 1 (SREBP1) and suppresses lipogenesis in cancer cells. Oncogene, 35(5), 642–650. https://doi.org/10.1038/onc.2015.123
Chen, Z., Yu, D., Owonikoko, T. K., Ramalingam, S. S., & Sun, S. Y. (2021). Induction of SREBP1 degradation coupled with suppression of SREBP1-mediated lipogenesis impacts the response of EGFR mutant NSCLC cells to osimertinib. Oncogene, 40(49), 6653–6665. https://doi.org/10.1038/s41388-021-02057-0
Bengoechea-Alonso, M. T., & Ericsson, J. (2009). A phosphorylation cascade controls the degradation of active SREBP1. Journal of Biological Chemistry, 284(9), 5885–5895. https://doi.org/10.1074/jbc.M807906200
Dong, Q., Giorgianni, F., Beranova-Giorgianni, S., Deng, X., O’Meally, R. N., Bridges, D., Park, E. A., Cole, R. N., Elam, M. B., & Raghow, R. (2015). Glycogen synthase kinase-3-mediated phosphorylation of serine 73 targets sterol response element binding protein-1c (SREBP-1c) for proteasomal degradation. Bioscience Reports, 36(1), e00284. https://doi.org/10.1042/BSR20150234
Cervello, M., Augello, G., Cusimano, A., Emma, M. R., Balasus, D., Azzolina, A., McCubrey, J. A., & Montalto, G. (2017). Pivotal roles of glycogen synthase-3 in hepatocellular carcinoma. Advances in Biological Regulation, 65, 59–76. https://doi.org/10.1016/j.jbior.2017.06.002
Shin, S., Wolgamott, L., Yu, Y., Blenis, J., & Yoon, S. O. (2011). Glycogen synthase kinase (GSK)-3 promotes p70 ribosomal protein S6 kinase (p70S6K) activity and cell proliferation. Proceedings of the National Academy of the United States of America, 108(47), E1204–E1213. https://doi.org/10.1073/pnas.1110195108
Brohée, L., Crémer, J., Colige, A., & Deroanne, C. (2021). Lipin-1, a versatile regulator of lipid homeostasis, is a potential target for fighting cancer. International Journal of Molecular Sciences, 22(9). https://doi.org/10.3390/ijms22094419
Shimizu, K., Fukushima, H., Ogura, K., Lien, E. C., Nihira, N. T., Zhang, J., North, B. J., Guo, A., Nagashima, K., Nakagawa, T., Hoshikawa, S., Watahiki, A., Okabe, K., Yamada, A., Toker, A., Asara, J. M., Fukumoto, S., Nakayama, K. I., Nakayama, K., et al. (2017). The SCFβ-TRCP E3 ubiquitin ligase complex targets Lipin1 for ubiquitination and degradation to promote hepatic lipogenesis. Science Signaling, 10(460). https://doi.org/10.1126/scisignal.aah4117
Ishimoto, K., Nakamura, H., Tachibana, K., Yamasaki, D., Ota, A., Hirano, K. I., Tanaka, T., Hamakubo, T., Sakai, J., Kodama, T., & Doi, T. (2009). Sterol-mediated regulation of human lipin 1 gene expression in hepatoblastoma cells. Journal of Biological Chemistry, 284(33), 22195–22205. https://doi.org/10.1074/jbc.M109.028753
Han, H.-S., Kim, S. G., Kim, Y. S., Jang, S.-H., Kwon, Y., Choi, D., Huh, T., Moon, E., Ahn, E., Seong, J. K., Kweon, H.-S., Hwang, G.-S., Lee, D. H., Cho, K. W., & Koo, S.-H. (2022). A novel role of CRTC2 in promoting nonalcoholic fatty liver disease. Molecular Metabolism, 55, 101402. https://doi.org/10.1016/j.molmet.2021.101402
Kavanagh, K. L., Jörnvall, H., Persson, B., & Oppermann, U. (2008). Medium- and short-chain dehydrogenase/reductase gene and protein families : The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cellular and Molecular Life Sciences, 65(24), 3895–3906. https://doi.org/10.1007/s00018-008-8588-y
Yu, X., Li, Z., & Wu, W. K. (2014). TIP30: A novel tumor-suppressor gene. Oncology research, 22(5-6), 339–348. https://doi.org/10.3727/096504015X14424348426116
Khemlina, G., Ikeda, S., & Kurzrock, R. (2017). The biology of hepatocellular carcinoma: Implications for genomic and immune therapies. Molecular Cancer, 16(1), 149. https://doi.org/10.1186/s12943-017-0712-x
Landras, A., Reger de Moura, C., Jouenne, F., Lebbe, C., Menashi, S., & Mourah, S. (2019). CD147 Is a promising target of tumor progression and a prognostic biomarker. Cancers, 11(11). https://doi.org/10.3390/cancers11111803
Cui, J., Huang, W., Wu, B., Jin, J., Jing, L., Shi, W. P., Liu, Z. Y., Yuan, L., Luo, D., Li, L., Chen, Z. N., & Jiang, J. L. (2018). N-Glycosylation by N-acetylglucosaminyltransferase V enhances the interaction of CD147/basigin with integrin β1 and promotes HCC metastasis. The Journal of Pathology, 245(1), 41–52. https://doi.org/10.1002/path.5054
Arvanitis, D., & Davy, A. (2008). Eph/ephrin signaling: Networks. Genes & Development, 22(4), 416–429. https://doi.org/10.1101/gad.1630408
Kullander, K., & Klein, R. (2002). Mechanisms and functions of Eph and ephrin signalling. Nature Reviews Molecular Cell Biology, 3(7), 475–486. https://doi.org/10.1038/nrm856
Pasquale, E. B. (2010). Eph receptors and ephrins in cancer: Bidirectional signalling and beyond. Nature Reviews Cancer, 10(3), 165–180. https://doi.org/10.1038/nrc2806
Iida, H., Honda, M., Kawai, H. F., Yamashita, T., Shirota, Y., Wang, B. C., Miao, H., & Kaneko, S. (2005). Ephrin-A1 expression contributes to the malignant characteristics of α-fetoprotein producing hepatocellular carcinoma. Gut, 54(6), 843. https://doi.org/10.1136/gut.2004.049486
Yuen, V. W., & Wong, C. C. (2020). Hypoxia-inducible factors and innate immunity in liver cancer. The Journal of Clinical Investigation, 130(10), 5052–5062. https://doi.org/10.1172/JCI137553
Sin, S. Q., Mohan, C. D., Goh, R. M. W., You, M., Nayak, S. C., Chen, L., Sethi, G., Rangappa, K. S., & Wang, L. (2022). Hypoxia signaling in hepatocellular carcinoma: Challenges and therapeutic opportunities. Cancer and Metastasis Reviews. https://doi.org/10.1007/s10555-022-10071-1
Li, Q., Yao, H., Wang, Y., Wu, Y., Thorne, R. F., Zhu, Y., Wu, M., & Liu, L. (2022). circPRKAA1 activates a Ku80/Ku70/SREBP-1 axis driving de novo fatty acid synthesis in cancer cells. Cell Reports, 41(8), 111707. https://doi.org/10.1016/j.celrep.2022.111707
Yang, J., Craddock, L., Hong, S., & Liu, Z. M. (2009). AMP-activated protein kinase suppresses LXR-dependent sterol regulatory element-binding protein-1c transcription in rat hepatoma McA-RH7777 cells. Journal of Cellular Biochemistry, 106(3), 414–426. https://doi.org/10.1002/jcb.22024
Yap, F., Craddock, L., & Yang, J. (2011). Mechanism of AMPK suppression of LXR-dependent Srebp-1c transcription. International Journal of Biological Sciences, 7(5), 645–650. https://doi.org/10.7150/ijbs.7.645
Wang, C., Tong, Y., Wen, Y., Cai, J., Guo, H., Huang, L., Xu, M., Feng, M., Chen, X., Zhang, J., Wu, H., Kong, X., & Xia, Q. (2018). Hepatocellular carcinoma-associated protein TD26 interacts and enhances sterol regulatory element-binding protein 1 activity to promote tumor cell proliferation and growth. Hepatology, 68(5), 1833–1850. https://doi.org/10.1002/hep.30030
Kohli, R. M., & Zhang, Y. (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature, 502(7472), 472–479. https://doi.org/10.1038/nature12750
Jansen, M., Ten Klooster, J. P., Offerhaus, G. J., & Clevers, H. (2009). LKB1 and AMPK family signaling: The intimate link between cell polarity and energy metabolism. Physiological Reviews, 89(3), 777–798. https://doi.org/10.1152/physrev.00026.2008
Jiang, X., Stockwell, B. R., & Conrad, M. (2021). Ferroptosis: Mechanisms, biology and role in disease. Nature Reviews Molecular Cell Biology, 22(4), 266–282. https://doi.org/10.1038/s41580-020-00324-8
Liberti, M. V., & Locasale, J. W. (2016). The Warburg effect: How does it benefit cancer cells? Trends in Biochemical Sciences, 41(3), 211–218. https://doi.org/10.1016/j.tibs.2015.12.001
Lee, H., Zandkarimi, F., Zhang, Y., Meena, J. K., Kim, J., Zhuang, L., Tyagi, S., Ma, L., Westbrook, T. F., Steinberg, G. R., Nakada, D., Stockwell, B. R., & Gan, B. (2020). Energy-stress-mediated AMPK activation inhibits ferroptosis. Nature Cell Biology, 22(2), 225–234. https://doi.org/10.1038/s41556-020-0461-8
Ates, G., Goldberg, J., Currais, A., & Maher, P. (2020). CMS121, a fatty acid synthase inhibitor, protects against excess lipid peroxidation and inflammation and alleviates cognitive loss in a transgenic mouse model of Alzheimer’s disease. Redox Biology, 36, 101648. https://doi.org/10.1016/j.redox.2020.101648
Bartolacci, C., Andreani, C., Vale, G., Berto, S., Melegari, M., Crouch, A. C., Baluya, D. L., Kemble, G., Hodges, K., Starrett, J., Politi, K., Starnes, S. L., Lorenzini, D., Raso, M. G., Solis Soto, L. M., Behrens, C., Kadara, H., Gao, B., Wistuba, I. I., et al. (2022). Targeting de novo lipogenesis and the Lands cycle induces ferroptosis in KRAS-mutant lung cancer. Nature Communications, 13(1), 4327. https://doi.org/10.1038/s41467-022-31963-4
Ye, Z., Zhuo, Q., Hu, Q., Xu, X., Mengqi, L., Zhang, Z., Xu, W., Liu, W., Fan, G., Qin, Y., Yu, X., & Ji, S. (2021). FBW7-NRA41-SCD1 axis synchronously regulates apoptosis and ferroptosis in pancreatic cancer cells. Redox Biology, 38, 101807. https://doi.org/10.1016/j.redox.2020.101807
Xuan, Y., Wang, H., Yung, M. M., Chen, F., Chan, W. S., Chan, Y. S., Tsui, S. K., Ngan, H. Y., Chan, K. K., & Chan, D. W. (2022). SCD1/FADS2 fatty acid desaturases equipoise lipid metabolic activity and redox-driven ferroptosis in ascites-derived ovarian cancer cells. Theranostics, 12(7), 3534–3552. https://doi.org/10.7150/thno.70194
Tesfay, L., Paul, B. T., Konstorum, A., Deng, Z., Cox, A. O., Lee, J., Furdui, C. M., Hegde, P., Torti, F. M., & Torti, S. V. (2019). Stearoyl-CoA desaturase 1 protects ovarian cancer cells from ferroptotic cell death. Cancer Research, 79(20), 5355–5366. https://doi.org/10.1158/0008-5472.CAN-19-0369
Carbone, M., & Melino, G. (2019). Stearoyl CoA desaturase regulates ferroptosis in ovarian cancer offering new therapeutic perspectives. Cancer Research, 79(20), 5149–5150. https://doi.org/10.1158/0008-5472.CAN-19-2453
Doll, S., Proneth, B., Tyurina, Y. Y., Panzilius, E., Kobayashi, S., Ingold, I., Irmler, M., Beckers, J., Aichler, M., Walch, A., Prokisch, H., Trümbach, D., Mao, G., Qu, F., Bayir, H., Füllekrug, J., Scheel, C. H., Wurst, W., Schick, J. A., et al. (2017). ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nature Chemical Biology, 13(1), 91–98. https://doi.org/10.1038/nchembio.2239
Li, C., Dong, X., Du, W., Shi, X., Chen, K., Zhang, W., & Gao, M. (2020). LKB1-AMPK axis negatively regulates ferroptosis by inhibiting fatty acid synthesis. Signal Transduction and Targeted Therapy, 5(1), 187. https://doi.org/10.1038/s41392-020-00297-2
Shi, Z., Liu, R., Lu, Q., Zeng, Z., Liu, Y., Zhao, J., Liu, X., Li, L., Huang, H., Yao, Y., Huang, D., & Xu, Q. (2021). UBE2O promotes hepatocellular carcinoma cell proliferation and invasion by regulating the AMPKalpha2/mTOR pathway. International Journal of Medical Sciences, 18(16), 3749–3758. https://doi.org/10.7150/ijms.63220
Wang, X., Li, Q., Sui, B., Xu, M., Pu, Z., & Qiu, T. (2022). Schisandrin A from Schisandra chinensis attenuates ferroptosis and NLRP3 inflammasome-mediated pyroptosis in diabetic nephropathy through mitochondrial damage by AdipoR1 ubiquitination. Oxidative Medicine and Cellular Longevity, 2022, 5411462. https://doi.org/10.1155/2022/5411462
Furman, D., Campisi, J., Verdin, E., Carrera-Bastos, P., Targ, S., Franceschi, C., Ferrucci, L., Gilroy, D. W., Fasano, A., Miller, G. W., Miller, A. H., Mantovani, A., Weyand, C. M., Barzilai, N., Goronzy, J. J., Rando, T. A., Effros, R. B., Lucia, A., Kleinstreuer, N., & Slavich, G. M. (2019). Chronic inflammation in the etiology of disease across the life span. Nature Medicine, 25(12), 1822–1832. https://doi.org/10.1038/s41591-019-0675-0
Ringelhan, M., Pfister, D., O’Connor, T., Pikarsky, E., & Heikenwalder, M. (2018). The immunology of hepatocellular carcinoma. Nature Immunology, 19(3), 222–232. https://doi.org/10.1038/s41590-018-0044-z
Karin, M., Lawrence, T., & Nizet, V. (2006). Innate immunity gone awry: Linking microbial infections to chronic inflammation and cancer. Cell, 124(4), 823–835. https://doi.org/10.1016/j.cell.2006.02.016
Budhu, A., & Wang, X. W. (2006). The role of cytokines in hepatocellular carcinoma. Journal of Leukocyte Biology, 80(6), 1197–1213. https://doi.org/10.1189/jlb.0506297
He, G., & Karin, M. (2011). NF-κB and STAT3 - Key players in liver inflammation and cancer. Cell Research, 21(1), 159–168. https://doi.org/10.1038/cr.2010.183
Greten, F. R., & Grivennikov, S. I. (2019). Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity, 51(1), 27–41. https://doi.org/10.1016/j.immuni.2019.06.025
Todoric, J., & Karin, M. (2019). The fire within: Cell-autonomous mechanisms in inflammation-driven cancer. Cancer Cell, 35(5), 714–720. https://doi.org/10.1016/j.ccell.2019.04.001
Lv, Q., Zhen, Q., Liu, L., Gao, R., Yang, S., Zhou, H., Goswami, R., & Li, Q. (2015). AMP-kinase pathway is involved in tumor necrosis factor alpha-induced lipid accumulation in human hepatoma cells. Life Sciences, 131, 23–29. https://doi.org/10.1016/j.lfs.2015.03.003
Kumar, D. P., Santhekadur, P. K., Seneshaw, M., Mirshahi, F., Uram-Tuculescu, C., & Sanyal, A. J. (2019). A regulatory role of apoptosis antagonizing transcription factor in the pathogenesis of nonalcoholic fatty liver disease and hepatocellular carcinoma. Hepatology, 69(4), 1520–1534. https://doi.org/10.1002/hep.30346
Shankaraiah, R. C., Callegari, E., Guerriero, P., Rimessi, A., Pinton, P., Gramantieri, L., Silini, E. M., Sabbioni, S., & Negrini, M. (2019). Metformin prevents liver tumourigenesis by attenuating fibrosis in a transgenic mouse model of hepatocellular carcinoma. Oncogene, 38(45), 7035–7045. https://doi.org/10.1038/s41388-019-0942-z
Wang, Y., Luo, M., Wang, F., Tong, Y., Li, L., Shu, Y., Qiao, K., Zhang, L., Yan, G., Liu, J., Ji, H., Xie, Y., Zhang, Y., Gao, W. Q., & Liu, Y. (2022). AMPK induces degradation of the transcriptional repressor PROX1 impairing branched amino acid metabolism and tumourigenesis. Nature Communications, 13(1), 7215. https://doi.org/10.1038/s41467-022-34747-y
Yang, X., Liu, Y., Li, M., Wu, H., Wang, Y., You, Y., Li, P., Ding, X., Liu, C., & Gong, J. (2018). Predictive and preventive significance of AMPK activation on hepatocarcinogenesis in patients with liver cirrhosis. Cell Death & Disease, 9(3), 264. https://doi.org/10.1038/s41419-018-0308-4
Chalasani, N., Vuppalanchi, R., Rinella, M., Middleton, M. S., Siddiqui, M. S., Barritt, A. S., Kolterman, O., Flores, O., Alonso, C., Iruarrizaga-Lejarreta, M., Gil-Redondo, R., Sirlin, C. B., & Zemel, M. B. (2018). Randomised clinical trial: a leucine-metformin-sildenafil combination (NS-0200) vs placebo in patients with non-alcoholic fatty liver disease. Alimentary Pharmacology and Therapeutics, 47(12), 1639–1651. https://doi.org/10.1111/apt.14674
Ma, T., Tian, X., Zhang, B., Li, M., Wang, Y., Yang, C., Wu, J., Wei, X., Qu, Q., Yu, Y., Long, S., Feng, J. W., Li, C., Zhang, C., Xie, C., Wu, Y., Xu, Z., Chen, J., Yu, Y., et al. (2022). Low-dose metformin targets the lysosomal AMPK pathway through PEN2. Nature, 603(7899), 159–165. https://doi.org/10.1038/s41586-022-04431-8
Wang, Y., An, H., Liu, T., Qin, C., Sesaki, H., Guo, S., Radovick, S., Hussain, M., Maheshwari, A., Wondisford, F. E., O’Rourke, B., & He, L. (2019). Metformin improves mitochondrial respiratory activity through activation of AMPK. Cell Reports, 29(6), 1511–1523.e1515. https://doi.org/10.1016/j.celrep.2019.09.070
Rena, G., Hardie, D. G., & Pearson, E. R. (2017). The mechanisms of action of metformin. Diabetologia, 60(9), 1577–1585. https://doi.org/10.1007/s00125-017-4342-z
Tobar, N., Rocha, G. Z., Santos, A., Guadagnini, D., Assalin, H. B., Camargo, J. A., Gonçalves, A., Pallis, F. R., Oliveira, A. G., Rocco, S. A., Neto, R. M., de Sousa, I. L., Alborghetti, M. R., Sforça, M. L., Rodrigues, P. B., Ludwig, R. G., Vanzela, E. C., Brunetto, S. Q., Boer, P. A., et al. (2023). Metformin acts in the gut and induces gut-liver crosstalk. Proceedings of the National Academy of Sciences of the United States of America, 120(4), e2211933120. https://doi.org/10.1073/pnas.2211933120
Pernicova, I., & Korbonits, M. (2014). Metformin--mode of action and clinical implications for diabetes and cancer. Nature Reviews Endocrinology, 10(3), 143–156. https://doi.org/10.1038/nrendo.2013.256
Kong, L., Zhang, H., Lu, C., Shi, K., Huang, H., Zheng, Y., Wang, Y., Wang, D., Wang, H., & Huang, W. (2021). AICAR, an AMP-activated protein kinase activator, ameliorates acute pancreatitis-associated liver injury partially through Nrf2-mediated antioxidant effects and inhibition of NLRP3 inflammasome activation. Frontiers in Pharmacology, 12, 724514. https://doi.org/10.3389/fphar.2021.724514
Gao, J., Xiong, R., Xiong, D., Zhao, W., Zhang, S., Yin, T., Zhang, X., Jiang, G., & Yin, Z. (2018). The adenosine monophosphate (AMP) analog, 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) inhibits hepatosteatosis and liver tumorigenesis in a high-fat diet murine model treated with diethylnitrosamine (DEN). Medical Science Monitor, 24, 8533–8543. https://doi.org/10.12659/msm.910544
Suski, M., Wiśniewska, A., Stachowicz, A., Olszanecki, R., Kuś, K., Białas, M., Madej, J., & Korbut, R. (2017). The influence of AICAR - direct activator of AMP-activated protein kinase (AMPK) - on liver proteome in apoE-knockout mice. European Journal of Pharmaceutical Sciences, 104, 406–416. https://doi.org/10.1016/j.ejps.2017.04.021
Miller, R. A., & Birnbaum, M. J. (2010). An energetic tale of AMPK-independent effects of metformin. The Journal of Clinical Investigation, 120(7), 2267–2270. https://doi.org/10.1172/jci43661
Foretz, M., Guigas, B., Bertrand, L., Pollak, M., & Viollet, B. (2014). Metformin: from mechanisms of action to therapies. Cell Metabolism, 20(6), 953–966. https://doi.org/10.1016/j.cmet.2014.09.018
Gores, G. J. (2014). Decade in review-hepatocellular carcinoma: HCC-subtypes, stratification and sorafenib. Nature Reviews Gastroenterology & Hepatology, 11(11), 645–647. https://doi.org/10.1038/nrgastro.2014.157
Tang, W., Chen, Z., Zhang, W., Cheng, Y., Zhang, B., Wu, F., Wang, Q., Wang, S., Rong, D., Reiter, F. P., De Toni, E. N., & Wang, X. (2020). The mechanisms of sorafenib resistance in hepatocellular carcinoma: Theoretical basis and therapeutic aspects. Signal Transduction and Targeted Therapy, 5(1), 87. https://doi.org/10.1038/s41392-020-0187-x
Inoki, K., Zhu, T., & Guan, K. L. (2003). TSC2 mediates cellular energy response to control cell growth and survival. Cell, 115(5), 577–590. https://doi.org/10.1016/S0092-8674(03)00929-2
Gwinn, D. M., Shackelford, D. B., Egan, D. F., Mihaylova, M. M., Mery, A., Vasquez, D. S., Turk, B. E., & Shaw, R. J. (2008). AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular Cell, 30(2), 214–226. https://doi.org/10.1016/j.molcel.2008.03.003
Van Nostrand, J. L., Hellberg, K., Luo, E. C., Van Nostrand, E. L., Dayn, A., Yu, J., Shokhirev, M. N., Dayn, Y., Yeo, G. W., & Shaw, R. J. (2020). AMPK regulation of Raptor and TSC2 mediate metformin effects on transcriptional control of anabolism and inflammation. Genes & Development, 34(19-20), 1330–1344. https://doi.org/10.1101/gad.339895.120
Shang, J., Chen, L.-L., Xiao, F.-X., Sun, H., Ding, H.-C., & Xiao, H. (2008). Resveratrol improves non-alcoholic fatty liver disease by activating AMP-activated protein kinase. Acta Pharmacologica Sinica, 29(6), 698–706. https://doi.org/10.1111/j.1745-7254.2008.00807.x
Chang, J. J., Hsu, M. J., Huang, H. P., Chung, D. J., Chang, Y. C., & Wang, C. J. (2013). Mulberry anthocyanins inhibit oleic acid induced lipid accumulation by reduction of lipogenesis and promotion of hepatic lipid clearance. Journal of Agricultural and Food Chemistry, 61(25), 6069–6076. https://doi.org/10.1021/jf401171k
Lee, M. S., Kim, J. S., Cho, S. M., Lee, S. O., Kim, S. H., & Lee, H. J. (2015). Fermented Rhus verniciflua stokes extract exerts an antihepatic lipogenic effect in oleic-acid-induced HepG2 cells via upregulation of AMP-activated protein kinase. Journal of Agricultural and Food Chemistry, 63(32), 7270–7276. https://doi.org/10.1021/acs.jafc.5b01954
Dong, Q., Giorgianni, F., Deng, X., Beranova-Giorgianni, S., Bridges, D., Park, E. A., Raghow, R., & Elam, M. B. (2014). Phosphorylation of sterol regulatory element binding protein-1a by protein kinase A (PKA) regulates transcriptional activity. Biochemical and Biophysical Research Communications, 449(4), 449–454. https://doi.org/10.1016/j.bbrc.2014.05.046
Ferretti, A. C., Tonucci, F. M., Hidalgo, F., Almada, E., Larocca, M. C., & Favre, C. (2016). AMPK and PKA interaction in the regulation of survival of liver cancer cells subjected to glucose starvation. Oncotarget, 7(14), 17815–17828. https://doi.org/10.18632/oncotarget.7404
Lu, M., & Shyy, J. Y. (2006). Sterol regulatory element-binding protein 1 is negatively modulated by PKA phosphorylation. American Journal of Physiology. Cell Physiology, 290(6), C1477–C1486. https://doi.org/10.1152/ajpcell.00374.2005
Lin, C. Y., & Gustafsson, J. (2015). Targeting liver X receptors in cancer therapeutics. Nature Reviews Cancer, 15(4), 216–224. https://doi.org/10.1038/nrc3912
Edwards, P. A., Kennedy, M. A., & Mak, P. A. (2002). LXRs;: Oxysterol-activated nuclear receptors that regulate genes controlling lipid homeostasis. Vascular Pharmacology, 38(4), 249–256. https://doi.org/10.1016/S1537-1891(02)00175-1
Steffensen, K. R., Gustafsson, J.-A., & k. (2004). Putative metabolic effects of the liver X. Receptor (LXR), 53(suppl_1), S36–S42.
Fernández-Alvarez, A., Alvarez, M. S., Gonzalez, R., Cucarella, C., Muntané, J., & Casado, M. (2011). Human SREBP1c expression in liver is directly regulated by peroxisome proliferator-activated receptor alpha (PPARalpha). Journal of Biological Chemistry, 286(24), 21466–21477. https://doi.org/10.1074/jbc.M110.209973
Zhou, Y., Yu, S., Cai, C., Zhong, L., Yu, H., & Shen, W. (2018). LXRɑ participates in the mTOR/S6K1/SREBP-1c signaling pathway during sodium palmitate-induced lipogenesis in HepG2 cells. Nutrition & Metabolism, 15, 31. https://doi.org/10.1186/s12986-018-0268-9
Ou, J., Tu, H., Shan, B., Luk, A., DeBose-Boyd, R. A., Bashmakov, Y., Goldstein, J. L., & Brown, M. S. (2001). Unsaturated fatty acids inhibit transcription of the sterol regulatory element-binding protein-1c (SREBP-1c) gene by antagonizing ligand-dependent activation of the LXR. Proceedings of the National Academy of Sciences of the United States of America, 98(11), 6027–6032. https://doi.org/10.1073/pnas.111138698
Yoshikawa, T., Shimano, H., Amemiya-Kudo, M., Yahagi, N., Hasty, A. H., Matsuzaka, T., Okazaki, H., Tamura, Y., Iizuka, Y., Ohashi, K., Osuga, J., Harada, K., Gotoda, T., Kimura, S., Ishibashi, S., & Yamada, N. (2001). Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Molecular and Cellular Biology, 21(9), 2991–3000. https://doi.org/10.1128/MCB.21.9.2991-3000.2001
Jakel, H., Nowak, M., Moitrot, E., Dehondt, H., Hum, D. W., Pennacchio, L. A., Fruchart-Najib, J., & Fruchart, J. C. (2004). The liver X receptor ligand T0901317 down-regulates APOA5 gene expression through activation of SREBP-1c. Journal of Biological Chemistry, 279(44), 45462–45469. https://doi.org/10.1074/jbc.M404744200
Båvner, A., Johansson, L., Toresson, G., Gustafsson, J. A., & Treuter, E. (2002). A transcriptional inhibitor targeted by the atypical orphan nuclear receptor SHP. EMBO Reports, 3(5), 478–484. https://doi.org/10.1093/embo-reports/kvf087
Monroy-Ramirez, H. C., Galicia-Moreno, M., Sandoval-Rodriguez, A., Meza-Rios, A., Santos, A., & Armendariz-Borunda, J. (2021). PPARs as metabolic sensors and therapeutic targets in liver diseases. Intertional Journal of Molecular Sciences, 22(15). https://doi.org/10.3390/ijms22158298
Sauer, S. (2015). Ligands for the nuclear peroxisome proliferator-activated receptor gamma. Trends in Pharmacological Sciences, 36(10), 688–704. https://doi.org/10.1016/j.tips.2015.06.010
Qin, X., Xie, X., Fan, Y., Tian, J., Guan, Y., Wang, X., Zhu, Y., & Wang, N. (2008). Peroxisome proliferator-activated receptor-delta induces insulin-induced gene-1 and suppresses hepatic lipogenesis in obese diabetic mice. Hepatology, 48(2), 432–441. https://doi.org/10.1002/hep.22334
Lee, J. H., Kang, H. S., Park, H. Y., Moon, Y. A., Kang, Y. N., Oh, B. C., Song, D. K., Bae, J. H., & Im, S. S. (2017). PPARα-dependent Insig2a overexpression inhibits SREBP-1c processing during fasting. Scientific Reports, 7(1), 9958. https://doi.org/10.1038/s41598-017-10523-7
Konig, B., Koch, A., Spielmann, J., Hilgenfeld, C., Hirche, F., Stangl, G. I., & Eder, K. (2009). Activation of PPARalpha and PPARgamma reduces triacylglycerol synthesis in rat hepatoma cells by reduction of nuclear SREBP-1. European Journal of Pharmacology, 605(1-3), 23–30. https://doi.org/10.1016/j.ejphar.2009.01.009
Oberkofler, H., Schraml, E., Krempler, F., & Patsch, W. (2004). Restoration of sterol-regulatory-element-binding protein-1c gene expression in HepG2 cells by peroxisome-proliferator-activated receptor-gamma co-activator-1alpha. Biochemical Journal, 381(Pt 2), 357–363. https://doi.org/10.1042/BJ20040173
Lin, J., Yang, R., Tarr, P. T., Wu, P. H., Handschin, C., Li, S., Yang, W., Pei, L., Uldry, M., Tontonoz, P., Newgard, C. B., & Spiegelman, B. M. (2005). Hyperlipidemic effects of dietary saturated fats mediated through PGC-1beta coactivation of SREBP. Cell, 120(2), 261–273. https://doi.org/10.1016/j.cell.2004.11.043
Hu, X., Li, J., Fu, M., Zhao, X., & Wang, W. (2021). The JAK/STAT signaling pathway: From bench to clinic. Signal Transduction and Targeted Therapy, 6(1), 402. https://doi.org/10.1038/s41392-021-00791-1
Athavale, D., Chouhan, S., Pandey, V., Mayengbam, S. S., Singh, S., & Bhat, M. K. (2018). Hepatocellular carcinoma-associated hypercholesterolemia: Involvement of proprotein-convertase-subtilisin-kexin type-9 (PCSK9). Cancer & Metabolism, 6, 16. https://doi.org/10.1186/s40170-018-0187-2
Sekiya, M., Hiraishi, A., Touyama, M., & Sakamoto, K. (2008). Oxidative stress induced lipid accumulation via SREBP1c activation in HepG2 cells. Biochemical and Biophysical Research Communications, 375(4), 602–607. https://doi.org/10.1016/j.bbrc.2008.08.068
Bosch-Barrera, J., Queralt, B., & Menendez, J. A. (2017). Targeting STAT3 with silibinin to improve cancer therapeutics. Cancer Treatment Reviews, 58, 61–69. https://doi.org/10.1016/j.ctrv.2017.06.003
Shi, Z., Zhou, Q., Gao, S., Li, W., Li, X., Liu, Z., Jin, P., & Jiang, J. (2019). Silibinin inhibits endometrial carcinoma via blocking pathways of STAT3 activation and SREBP1-mediated lipid accumulation. Life Sciences, 217, 70–80. https://doi.org/10.1016/j.lfs.2018.11.037
Wang, C., Zhang, J., Yin, J., Gan, Y., Xu, S., Gu, Y., & Huang, W. (2021). Alternative approaches to target Myc for cancer treatment. Signal Transduction and Targeted Therapy, 6(1), 117. https://doi.org/10.1038/s41392-021-00500-y
Dong, Y., Tu, R., Liu, H., & Qing, G. (2020). Regulation of cancer cell metabolism: oncogenic MYC in the driver’s seat. Signal Transduction and Targeted Therapy, 5(1), 124. https://doi.org/10.1038/s41392-020-00235-2
Chen, J., Ding, C., Chen, Y., Hu, W., Yu, C., Peng, C., Feng, X., Cheng, Q., Wu, W., Lu, Y., Xie, H., Zhou, L., Wu, J., & Zheng, S. (2021). ACSL4 reprograms fatty acid metabolism in hepatocellular carcinoma via c-Myc/SREBP1 pathway. Cancer Letters, 502, 154–165. https://doi.org/10.1016/j.canlet.2020.12.019
Carroll, P. A., Diolaiti, D., McFerrin, L., Gu, H., Djukovic, D., Du, J., Cheng, P. F., Anderson, S., Ulrich, M., Hurley, J. B., Raftery, D., Ayer, D. E., & Eisenman, R. N. (2015). Deregulated Myc requires MondoA/Mlx for metabolic reprogramming and tumorigenesis. Cancer Cell, 27(2), 271–285. https://doi.org/10.1016/j.ccell.2014.11.024
Chen, J., Ding, C., Chen, Y., Hu, W., Lu, Y., Wu, W., Zhang, Y., Yang, B., Wu, H., Peng, C., Xie, H., Zhou, L., Wu, J., & Zheng, S. (2020). ACSL4 promotes hepatocellular carcinoma progression via c-Myc stability mediated by ERK/FBW7/c-Myc axis. Oncogenesis, 9(4), 42. https://doi.org/10.1038/s41389-020-0226-z
Bengoechea-Alonso, M. T., & Ericsson, J. (2006). Cdk1/cyclin B-mediated phosphorylation stabilizes SREBP1 during mitosis. Cell Cycle, 5(15), 1708–1718. https://doi.org/10.4161/cc.5.15.3131
Asghar, U., Witkiewicz, A. K., Turner, N. C., & Knudsen, E. S. (2015). The history and future of targeting cyclin-dependent kinases in cancer therapy. Nature Reviews Drug Discovery, 14(2), 130–146. https://doi.org/10.1038/nrd4504
Xu, Q., Liu, X., Zheng, X., Yao, Y., & Liu, Q. (2014). PKM2 regulates Gli1 expression in hepatocellular carcinoma. Oncology Letters, 8(5), 1973–1979. https://doi.org/10.3892/ol.2014.2441
Hacker, H. J., Steinberg, P., & Bannasch, P. (1998). Pyruvate kinase isoenzyme shift from L-type to M2-type is a late event in hepatocarcinogenesis induced in rats by a choline-deficient/DL-ethionine-supplemented diet. Carcinogenesis, 19(1), 99–107. https://doi.org/10.1093/carcin/19.1.99
Tani, K., Yoshida, M. C., Satoh, H., Mitamura, K., Noguchi, T., Tanaka, T., Fujii, H., & Miwa, S. (1988). Human M2-type pyruvate kinase: cDNA cloning, chromosomal assignment and expression in hepatoma. Gene, 73(2), 509–516. https://doi.org/10.1016/0378-1119(88)90515-X
Reinacher, M., Eigenbrodt, E., Gerbracht, U., Zenk, G., Timmermann-Trosiener, I., Bentley, P., Waechter, F., & Schulte-Hermann, R. (1986). Pyruvate kinase isoenzymes in altered foci and carcinoma of rat liver. Carcinogenesis, 7(8), 1351–1357. https://doi.org/10.1093/carcin/7.8.1351
Alquraishi, M., Puckett, D. L., Alani, D. S., Humidat, A. S., Frankel, V. D., Donohoe, D. R., Whelan, J., & Bettaieb, A. (2019). Pyruvate kinase M2: A simple molecule with complex functions. Free Radical Biology and Medicine, 143, 176–192. https://doi.org/10.1016/j.freeradbiomed.2019.08.007
Méndez-Lucas, A., Li, X., Hu, J., Che, L., Song, X., Jia, J., Wang, J., Xie, C., Driscoll, P. C., Tschaharganeh, D. F., Calvisi, D. F., Yuneva, M., & Chen, X. (2017). Glucose catabolism in liver tumors induced by c-MYC can be sustained by various PKM1/PKM2 ratios and pyruvate kinase activities. Cancer Research, 77(16), 4355–4364. https://doi.org/10.1158/0008-5472.CAN-17-0498
Giandomenico, V., Simonsson, M., Gronroos, E., & Ericsson, J. (2003). Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Molecular and Cellular Biology, 23(7), 2587–2599. https://doi.org/10.1128/MCB.23.7.2587-2599.2003
Wu, D., Yang, Y., Hou, Y., Zhao, Z., Liang, N., Yuan, P., Yang, T., Xing, J., & Li, J. (2022). Increased mitochondrial fission drives the reprogramming of fatty acid metabolism in hepatocellular carcinoma cells through suppression of Sirtuin 1. Cancer Communications, 42(1), 37–55. https://doi.org/10.1002/cac2.12247
Wu, Q. J., Zhang, T. N., Chen, H. H., Yu, X. F., Lv, J. L., Liu, Y. Y., Liu, Y. S., Zheng, G., Zhao, J. Q., Wei, Y. F., Guo, J. Y., Liu, F. H., Chang, Q., Zhang, Y. X., Liu, C. G., & Zhao, Y. H. (2022). The sirtuin family in health and disease. Signal Transduction and Targeted Therapy, 7(1), 402. https://doi.org/10.1038/s41392-022-01257-8
Alves-Fernandes, D. K., & Jasiulionis, M. G. (2019). The role of SIRT1 on DNA damage response and epigenetic alterations in cancer. International Journal of Molecular Sciences, 20(13). https://doi.org/10.3390/ijms20133153
Ren, R., Wang, Z., Wu, M., & Wang, H. (2020). Emerging roles of SIRT1 in alcoholic liver disease. International Journal of Biological Sciences, 16(16), 3174–3183. https://doi.org/10.7150/ijbs.49535
Quintana-Cabrera, R., & Scorrano, L. (2023). Determinants and outcomes of mitochondrial dynamics. Molecular Cell, 83(6), 857–876. https://doi.org/10.1016/j.molcel.2023.02.012
Geisbrecht, B. V., & Gould, S. J. (1999). The human PICD gene encodes a cytoplasmic and peroxisomal NADP(+)-dependent isocitrate dehydrogenase. Journal of Biological Chemistry, 274(43), 30527–30533. https://doi.org/10.1074/jbc.274.43.30527
D'Adamo, A. F., Jr., & Haft, D. E. (1965). An alternate pathway of alpha-ketoglutarate catabolism in the isolated, perfused rat liver. I. Studies with Dl-glutamate-2- and -5-14c. Journal of Biological Chemistry, 240, 613–617. https://doi.org/10.1016/S0021-9258(17)45218-5
Ochoa, S., & Weisz-Tabori, E. (1948). Biosynthesis of tricarboxylic acids by carbon dioxide fixation; oxalosuccinic carboxylase. Journal of Biological Chemistry, 174(1), 123–132. https://doi.org/10.1016/S0021-9258(18)57382-8
Filipp, F. V., Scott, D. A., Ronai, Z. A., Osterman, A. L., & Smith, J. W. (2012). Reverse TCA cycle flux through isocitrate dehydrogenases 1 and 2 is required for lipogenesis in hypoxic melanoma cells. Pigment Cell & Melanoma, 25(3), 375–383. https://doi.org/10.1111/j.1755-148X.2012.00989.x
Metallo, C. M., Gameiro, P. A., Bell, E. L., Mattaini, K. R., Yang, J., Hiller, K., Jewell, C. M., Johnson, Z. R., Irvine, D. J., Guarente, L., Kelleher, J. K., Vander Heiden, M. G., Iliopoulos, O., & Stephanopoulos, G. (2011). Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature, 481(7381), 380–384. https://doi.org/10.1038/nature10602
Liu, L., Zhao, X., Zhao, L., Li, J., Yang, H., Zhu, Z., Liu, J., & Huang, G. (2016). Arginine methylation of SREBP1a via PRMT5 promotes de novo lipogenesis and tumor growth. Cancer Research, 76(5), 1260–1272. https://doi.org/10.1158/0008-5472.CAN-15-1766
Enchev, R. I., Schulman, B. A., & Peter, M. (2015). Protein neddylation: beyond cullin-RING ligases. Nature Reviews Molecular Cell Biology, 16(1), 30–44. https://doi.org/10.1038/nrm3919
Tai, Y., Gao, J. H., Zhao, C., Tong, H., Zheng, S. P., Huang, Z. Y., Liu, R., Tang, C. W., & Li, J. (2018). SK-Hep1: Not hepatocellular carcinoma cells but a cell model for liver sinusoidal endothelial cells. International Journal of Clinical & Experimental Pathology, 11(5), 2931–2938.
Brown, M. S., & Goldstein, J. L. (1997). The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell, 89(3), 331–340. https://doi.org/10.1016/S0092-8674(00)80213-5
Wang, S. L., Du, E. Z., Martin, T. D., & Davis, R. A. (1997). Coordinate regulation of lipogenesis, the assembly and secretion of apolipoprotein B-containing lipoproteins by sterol response element binding protein 1. Journal of Biological Chemistry, 272(31), 19351–19358. https://doi.org/10.1074/jbc.272.31.19351
Ren, S., & Ning, Y. (2014). Sulfation of 25-hydroxycholesterol regulates lipid metabolism, inflammatory responses, and cell proliferation. American Journal of Physiology. Endocrinology and Metabolism, 306(2), E123–E130. https://doi.org/10.1152/ajpendo.00552.2013
Peng, W., Liu, C., Xu, C., Lou, Y., Chen, J., Yang, Y., Yagita, H., Overwijk, W. W., Lizée, G., Radvanyi, L., & Hwu, P. (2012). PD-1 blockade enhances T-cell migration to tumors by elevating IFN-γ inducible chemokines. Cancer Research, 72(20), 5209–5218.
Norlin, M., & Chiang, J. Y. (2004). Transcriptional regulation of human oxysterol 7alpha-hydroxylase by sterol response element binding protein. Biochemical and Biophysical Research Communications, 316(1), 158–164. https://doi.org/10.1016/j.bbrc.2004.02.029
Beigneux, A., Hofmann, A. F., & Young, S. G. (2002). Human CYP7A1 deficiency: Progress and enigmas. The Journal of Clinical Investigation, 110(1), 29–31. https://doi.org/10.1172/JCI16076
Botolin, D., Wang, Y., Christian, B., & Jump, D. B. (2006). Docosahexaneoic acid (22:6,n-3) regulates rat hepatocyte SREBP-1 nuclear abundance by Erk- and 26S proteasome-dependent pathways. Journal of Lipid Research, 47(1), 181–192. https://doi.org/10.1194/jlr.M500365-JLR200
Guillou, H., Zadravec, D., Martin, P. G., & Jacobsson, A. (2010). The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Progress in Lipid Research, 49(2), 186–199. https://doi.org/10.1016/j.plipres.2009.12.002
Sawada, N., Inoue, M., Iwasaki, M., Sasazuki, S., Shimazu, T., Yamaji, T., Takachi, R., Tanaka, Y., Mizokami, M., & Tsugane, S. (2012). Consumption of n-3 fatty acids and fish reduces risk of hepatocellular carcinoma. Gastroenterology, 142(7), 1468–1475. https://doi.org/10.1053/j.gastro.2012.02.018
Koh, W. P., Dan, Y. Y., Goh, G. B., Jin, A., Wang, R., & Yuan, J. M. (2016). Dietary fatty acids and risk of hepatocellular carcinoma in the Singapore Chinese health study. Liver International, 36(6), 893–901. https://doi.org/10.1111/liv.12978
Nanthirudjanar, T., Furumoto, H., Hirata, T., & Sugawara, T. (2013). Oxidized eicosapentaenoic acids more potently reduce LXRalpha-induced cellular triacylglycerol via suppression of SREBP-1c, PGC-1beta and GPA than its intact form. Lipids in Health and Disease, 12, 73. https://doi.org/10.1186/1476-511X-12-73
Koeberle, A., & Werz, O. (2018). Natural products as inhibitors of prostaglandin E(2) and pro-inflammatory 5-lipoxygenase-derived lipid mediator biosynthesis. Biotechnology Advances, 36(6), 1709–1723. https://doi.org/10.1016/j.biotechadv.2018.02.010
Chen, J., Li, X., Ge, C., Min, J., & Wang, F. (2022). The multifaceted role of ferroptosis in liver disease. Cell Death & Differentiation, 29(3), 467–480. https://doi.org/10.1038/s41418-022-00941-0
Rong, X., Wang, B., Palladino, E. N., de Aguiar Vallim, T. Q., Ford, D. A., & Tontonoz, P. (2017). ER phospholipid composition modulates lipogenesis during feeding and in obesity. The Journal of Clinical Investigation, 127(10), 3640–3651. https://doi.org/10.1172/jci93616
Wang, B., & Tontonoz, P. (2019). Phospholipid remodeling in physiology and disease. Annual Review of Physiology, 81, 165–188. https://doi.org/10.1146/annurev-physiol-020518-114444
Valentine, W. J., Yanagida, K., Kawana, H., Kono, N., Noda, N. N., Aoki, J., & Shindou, H. (2022). Update and nomenclature proposal for mammalian lysophospholipid acyltransferases, which create membrane phospholipid diversity. Journal of Biological Chemistry, 298(1), 101470. https://doi.org/10.1016/j.jbc.2021.101470
Dobrosotskaya, I. Y., Seegmiller, A. C., Brown, M. S., Goldstein, J. L., & Rawson, R. B. (2002). Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science, 296(5569), 879–883. https://doi.org/10.1126/science.1071124
Walker, A. K., Jacobs, R. L., Watts, J. L., Rottiers, V., Jiang, K., Finnegan, D. M., Shioda, T., Hansen, M., Yang, F., Niebergall, L. J., Vance, D. E., Tzoneva, M., Hart, A. C., & Näär, A. M. (2011). A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell, 147(4), 840–852. https://doi.org/10.1016/j.cell.2011.09.045
Koeberle, A., Shindou, H., Koeberle, S. C., Laufer, S. A., Shimizu, T., & Werz, O. (2013). Arachidonoyl-phosphatidylcholine oscillates during the cell cycle and counteracts proliferation by suppressing Akt membrane binding. Proceedings of the National Academy of Sciences of the United States of America, 110(7), 2546–2551. https://doi.org/10.1073/pnas.1216182110
Pein, H., Koeberle, S. C., Voelkel, M., Schneider, F., Rossi, A., Thürmer, M., Loeser, K., Sautebin, L., Morrison, H., Werz, O., & Koeberle, A. (2017). Vitamin A regulates Akt signaling through the phospholipid fatty acid composition. The FASEB Journal, 31(10), 4566–4577. https://doi.org/10.1096/fj.201700078R
Li, Y., Guo, D., Lu, G., Mohiuddin Chowdhury, A. T. M., Zhang, D., Ren, M., Chen, Y., Wang, R., & He, S. (2020). LncRNA SNAI3-AS1 promotes PEG10-mediated proliferation and metastasis via decoying of miR-27a-3p and miR-34a-5p in hepatocellular carcinoma. Cell Death & Disease, 11(8), 685. https://doi.org/10.1038/s41419-020-02840-z
Sun, Y., Xiao, Y., Sun, H., Zhao, Z., Zhu, J., Zhang, L., Dong, J., Han, T., Jing, Q., Zhou, J., & Jing, Z. (2019). miR-27a regulates vascular remodeling by targeting endothelial cells’ apoptosis and interaction with vascular smooth muscle cells in aortic dissection. Theranostics, 9(25), 7961–7975. https://doi.org/10.7150/thno.35737
Musto, A., Navarra, A., Vocca, A., Gargiulo, A., Minopoli, G., Romano, S., Romano, M. F., Russo, T., & Parisi, S. (2015). miR-23a, miR-24 and miR-27a protect differentiating ESCs from BMP4-induced apoptosis. Cell Death & Differentiation, 22(6), 1047–1057. https://doi.org/10.1038/cdd.2014.198
Tan, W., Zhang, Y., Li, M., Zhu, X., Yang, X., Wang, J., Zhang, S., Zhu, W., Cao, J., Yang, H., & Zhang, L. (2019). miR-27a-containing exosomes secreted by irradiated skin keratinocytes delayed the migration of unirradiated skin fibroblasts. International Journal of Biological Sciences, 15(10), 2240–2255. https://doi.org/10.7150/ijbs.35356
Towers, C. G., Guarnieri, A. L., Micalizzi, D. S., Harrell, J. C., Gillen, A. E., Kim, J., Wang, C. A., Oliphant, M. U. J., Drasin, D. J., Guney, M. A., Kabos, P., Sartorius, C. A., Tan, A. C., Perou, C. M., Espinosa, J. M., & Ford, H. L. (2015). The Six1 oncoprotein downregulates p53 via concomitant regulation of RPL26 and microRNA-27a-3p. Nature Communications, 6, 10077. https://doi.org/10.1038/ncomms10077
Shirasaki, T., Honda, M., Shimakami, T., Horii, R., Yamashita, T., Sakai, Y., Sakai, A., Okada, H., Watanabe, R., Murakami, S., Yi, M., Lemon, S. M., & Kaneko, S. (2013). MicroRNA-27a regulates lipid metabolism and inhibits hepatitis C virus replication in human hepatoma cells. Journal of Virology, 87(9), 5270–5286. https://doi.org/10.1128/jvi.03022-12
Viswanathan, S. R., Powers, J. T., Einhorn, W., Hoshida, Y., Ng, T. L., Toffanin, S., O’Sullivan, M., Lu, J., Phillips, L. A., Lockhart, V. L., Shah, S. P., Tanwar, P. S., Mermel, C. H., Beroukhim, R., Azam, M., Teixeira, J., Meyerson, M., Hughes, T. P., Llovet, J. M., et al. (2009). Lin28 promotes transformation and is associated with advanced human malignancies. Nature Genetics, 41(7), 843–848. https://doi.org/10.1038/ng.392
Wilbert, M. L., Huelga, S. C., Kapeli, K., Stark, T. J., Liang, T. Y., Chen, S. X., Yan, B. Y., Nathanson, J. L., Hutt, K. R., Lovci, M. T., Kazan, H., Vu, A. Q., Massirer, K. B., Morris, Q., Hoon, S., & Yeo, G. W. (2012). LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Molecular Cell, 48(2), 195–206. https://doi.org/10.1016/j.molcel.2012.08.004
Zhu, H., Shyh-Chang, N., Segrè, A. V., Shinoda, G., Shah, S. P., Einhorn, W. S., Takeuchi, A., Engreitz, J. M., Hagan, J. P., Kharas, M. G., Urbach, A., Thornton, J. E., Triboulet, R., Gregory, R. I., Altshuler, D., & Daley, G. Q. (2011). The Lin28/let-7 axis regulates glucose metabolism. Cell, 147(1), 81–94. https://doi.org/10.1016/j.cell.2011.08.033
Peng, S., Chen, L. L., Lei, X. X., Yang, L., Lin, H., Carmichael, G. G., & Huang, Y. (2011). Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells, 29(3), 496–504. https://doi.org/10.1002/stem.591
Li, N., Zhong, X., Lin, X., Guo, J., Zou, L., Tanyi, J. L., Shao, Z., Liang, S., Wang, L. P., Hwang, W. T., Katsaros, D., Montone, K., Zhao, X., & Zhang, L. (2012). Lin-28 homologue A (LIN28A) promotes cell cycle progression via regulation of cyclin-dependent kinase 2 (CDK2), cyclin D1 (CCND1), and cell division cycle 25 homolog A (CDC25A) expression in cancer. Journal of Biological Chemistry, 287(21), 17386–17397. https://doi.org/10.1074/jbc.M111.321158
Shyh-Chang, N., Zhu, H., Yvanka de Soysa, T., Shinoda, G., Seligson, M. T., Tsanov, K. M., Nguyen, L., Asara, J. M., Cantley, L. C., & Daley, G. Q. (2013). Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell, 155(4), 778–792. https://doi.org/10.1016/j.cell.2013.09.059
Thürmer, M., Gollowitzer, A., Pein, H., Neukirch, K., Gelmez, E., Waltl, L., Wielsch, N., Winkler, R., Löser, K., Grander, J., Hotze, M., Harder, S., Döding, A., Meßner, M., Troisi, F., Ardelt, M., Schlüter, H., Pachmayr, J., Gutiérrez-Gutiérrez, Ó., et al. (2022). PI(18:1/18:1) is a SCD1-derived lipokine that limits stress signaling. Nature Communications, 13(1), 2982. https://doi.org/10.1038/s41467-022-30374-9
Yang, Y., Ma, Y., Gao, H., Peng, T., Shi, H., Tang, Y., Li, H., Chen, L., Hu, K., & Han, A. (2021). A novel HDGF-ALCAM axis promotes the metastasis of Ewing sarcoma via regulating the GTPases signaling pathway. Oncogene, 40(4), 731–745. https://doi.org/10.1038/s41388-020-01485-8
Baude, A., Aaes, T. L., Zhai, B., Al-Nakouzi, N., Oo, H. Z., Daugaard, M., Rohde, M., & Jäättelä, M. (2016). Hepatoma-derived growth factor-related protein 2 promotes DNA repair by homologous recombination. Nucleic Acids Research, 44(5), 2214–2226. https://doi.org/10.1093/nar/gkv1526
Yang, Y., Li, H., Zhang, F., Shi, H., Zhen, T., Dai, S., Kang, L., Liang, Y., Wang, J., & Han, A. (2013). Clinical and biological significance of hepatoma-derived growth factor in Ewing’s sarcoma. The Journal of Pathology, 231(3), 323–334. https://doi.org/10.1002/path.4241
Chen, S. C., Kung, M. L., Hu, T. H., Chen, H. Y., Wu, J. C., Kuo, H. M., Tsai, H. E., Lin, Y. W., Wen, Z. H., Liu, J. K., Yeh, M. H., & Tai, M. H. (2012). Hepatoma-derived growth factor regulates breast cancer cell invasion by modulating epithelial--mesenchymal transition. The Journal of Pathology, 228(2), 158–169. https://doi.org/10.1002/path.3988
Everett, A. D., Lobe, D. R., Matsumura, M. E., Nakamura, H., & McNamara, C. A. (2000). Hepatoma-derived growth factor stimulates smooth muscle cell growth and is expressed in vascular development. The Journal of Clinical Investigation, 105(5), 567–575. https://doi.org/10.1172/JCI7497
Zhang, Y., Mohibi, S., Vasilatis, D. M., Chen, M., Zhang, J., & Chen, X. (2022). Ferredoxin reductase and p53 are necessary for lipid homeostasis and tumor suppression through the ABCA1-SREBP pathway. Oncogene, 41(12), 1718–1726. https://doi.org/10.1038/s41388-021-02100-0
Moon, S. H., Huang, C. H., Houlihan, S. L., Regunath, K., Freed-Pastor, W. A., Morris, J. P., Tschaharganeh, D. F., Kastenhuber, E. R., Barsotti, A. M., Culp-Hill, R., Xue, W., Ho, Y. J., Baslan, T., Li, X., Mayle, A., de Stanchina, E., Zender, L., Tong, D. R., D’Alessandro, A., et al. (2019). p53 represses the mevalonate pathway to mediate tumor suppression. Cell, 176(3), 564–580.e519. https://doi.org/10.1016/j.cell.2018.11.011
Mann, M. J., & Dzau, V. J. (2000). Therapeutic applications of transcription factor decoy oligonucleotides. The Journal of Clinical Investigation, 106(9), 1071–1075. https://doi.org/10.1172/jci11459
Kuzmich, N., Andresyuk, E., Porozov, Y., Tarasov, V., Samsonov, M., Preferanskaya, N., Veselov, V., & Alyautdin, R. (2022). PCSK9 as a target for development of a new generation of hypolipidemic drugs. Molecules, 27(2). https://doi.org/10.3390/molecules27020434
Shao, W., Machamer, C. E., & Espenshade, P. J. (2016). Fatostatin blocks ER exit of SCAP but inhibits cell growth in a SCAP-independent manner. Journal of Lipid Research, 57(8), 1564–1573. https://doi.org/10.1194/jlr.M069583
Ma, X., Zhao, T., Yan, H., Guo, K., Liu, Z., Wei, L., Lu, W., Qiu, C., & Jiang, J. (2021). Fatostatin reverses progesterone resistance by inhibiting the SREBP1-NF-κB pathway in endometrial carcinoma. Cell Death & Disease, 12(6), 544. https://doi.org/10.1038/s41419-021-03762-0
Gholkar, A. A., Cheung, K., Williams, K. J., Lo, Y. C., Hamideh, S. A., Nnebe, C., Khuu, C., Bensinger, S. J., & Torres, J. Z. (2016). Fatostatin inhibits cancer cell proliferation by affecting mitotic microtubule spindle assembly and cell division. Journal of Biological Chemistry, 291(33), 17001–17008. https://doi.org/10.1074/jbc.C116.737346
Li, X., Chen, Y. T., Hu, P., & Huang, W. C. (2014). Fatostatin displays high antitumor activity in prostate cancer by blocking SREBP-regulated metabolic pathways and androgen receptor signaling. Molecular Cancer Therapeutics, 13(4), 855–866. https://doi.org/10.1158/1535-7163.MCT-13-0797
Amiri, S., Dastghaib, S., Ahmadi, M., Mehrbod, P., Khadem, F., Behrouj, H., Aghanoori, M. R., Machaj, F., Ghamsari, M., Rosik, J., Hudecki, A., Afkhami, A., Hashemi, M., Los, M. J., Mokarram, P., Madrakian, T., & Ghavami, S. (2020). Betulin and its derivatives as novel compounds with different pharmacological effects. Biotechnology Advances, 38, 107409. https://doi.org/10.1016/j.biotechadv.2019.06.008
Guo, N., Miao, Y., & Sun, M. (2018). Transcatheter hepatic arterial chemoembolization plus cinobufotalin injection adjuvant therapy for advanced hepatocellular carcinoma: a meta-analysis of 27 trials involving 2,079 patients. OncoTargets and therapy, 11, 8835–8853. https://doi.org/10.2147/OTT.S182840
Li, H., Xiang, L., Yang, N., Cao, F., Li, C., Chen, P., Ruan, X., Feng, Y., Zhou, N., & Wang, X. (2018). Zhiheshouwu ethanol extract induces intrinsic apoptosis and reduces unsaturated fatty acids via SREBP1 pathway in hepatocellular carcinoma cells. Food and Chemical Toxicology, 119, 169–175. https://doi.org/10.1016/j.fct.2018.04.054
Le, T. N. H., Choi, H. J., & Jun, H. S. (2021). Ethanol Extract of Liriope platyphylla root attenuates non-alcoholic fatty liver disease in high-fat diet-induced obese mice via regulation of lipogenesis and lipid uptake. Nutrients, 13(10). https://doi.org/10.3390/nu13103338
Gong, X., Qian, H., Shao, W., Li, J., Wu, J., Liu, J. J., Li, W., Wang, H. W., Espenshade, P., & Yan, N. (2016). Complex structure of the fission yeast SREBP-SCAP binding domains reveals an oligomeric organization. Cell Research, 26(11), 1197–1211. https://doi.org/10.1038/cr.2016.123
Párraga, A., Bellsolell, L., Ferré-D’Amaré, A. R., & Burley, S. K. (1998). Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution. Structure, 6(5), 661–672. https://doi.org/10.1016/S0969-2126(98)00067-7
Yokoyama, C., Wang, X., Briggs, M. R., Admon, A., Wu, J., Hua, X., Goldstein, J. L., & Brown, M. S. (1993). SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell, 75(1), 187–197. https://doi.org/10.1016/S0092-8674(05)80095-9
Chen, R., Li, Q., Xu, S., Ye, C., Tian, T., Jiang, Q., Shan, J., & Ruan, J. (2022). Modulation of the tumour microenvironment in hepatocellular carcinoma by tyrosine kinase inhibitors: from modulation to combination therapy targeting the microenvironment. Cancer Cell International, 22(1), 73. https://doi.org/10.1186/s12935-021-02435-4
Forner, A., Reig, M., & Bruix, J. (2018). Hepatocellular carcinoma. Lancet, 391(10127), 1301–1314.
Howells, L. M., Berry, D. P., Elliott, P. J., Jacobson, E. W., Hoffmann, E., Hegarty, B., Brown, K., Steward, W. P., & Gescher, A. J. (2011). Phase I randomized, double-blind pilot study of micronized resveratrol (SRT501) in patients with hepatic metastases--safety, pharmacokinetics, and pharmacodynamics. Cancer Prevention Research, 4(9), 1419–1425. https://doi.org/10.1158/1940-6207.Capr-11-0148
Batchuluun, B., Pinkosky, S. L., & Steinberg, G. R. (2022). Lipogenesis inhibitors: Therapeutic opportunities and challenges. Nature Reviews Drug Discovery, 21(4), 283–305. https://doi.org/10.1038/s41573-021-00367-2
Kubota, C. S., & Espenshade, P. J. (2022). Targeting stearoyl-CoA desaturase in solid tumors. Cancer Research, 82(9), 1682–1688. https://doi.org/10.1158/0008-5472.CAN-21-4044
Feng, X., Zhang, L., Xu, S., & Shen, A. Z. (2020). ATP-citrate lyase (ACLY) in lipid metabolism and atherosclerosis: An updated review. Progress in Lipid Research, 77, 101006. https://doi.org/10.1016/j.plipres.2019.101006
Kong, F. H., Ye, Q. F., Miao, X. Y., Liu, X., Huang, S. Q., Xiong, L., Wen, Y., & Zhang, Z. J. (2021). Current status of sorafenib nanoparticle delivery systems in the treatment of hepatocellular carcinoma. Theranostics, 11(11), 5464–5490. https://doi.org/10.7150/thno.54822
Yang, S., Cai, C., Wang, H., Ma, X., Shao, A., Sheng, J., & Yu, C. (2022). Drug delivery strategy in hepatocellular carcinoma therapy. Cell Communication and Signaling, 20(1), 26. https://doi.org/10.1186/s12964-021-00796-x
Dutta, R., & Mahato, R. I. (2017). Recent advances in hepatocellular carcinoma therapy. Pharmacology & Therapeutics, 173, 106–117. https://doi.org/10.1016/j.pharmthera.2017.02.010
Naldini, L. (2015). Gene therapy returns to centre stage. Nature, 526(7573), 351–360. https://doi.org/10.1038/nature15818
Vandereyken, K., Sifrim, A., Thienpont, B., & Voet, T. (2023). Methods and applications for single-cell and spatial multi-omics. Nature Reviews Genetics, 1-22. https://doi.org/10.1038/s41576-023-00580-2
Gao, Q., Zhu, H., Dong, L., Shi, W., Chen, R., Song, Z., Huang, C., Li, J., Dong, X., Zhou, Y., Liu, Q., Ma, L., Wang, X., Zhou, J., Liu, Y., Boja, E., Robles, A. I., Ma, W., Wang, P., et al. (2019). Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. cell, 179(2), 561–577.e522. https://doi.org/10.1016/j.cell.2019.10.038
Ericksen, R. E., Lim, S. L., McDonnell, E., Shuen, W. H., Vadiveloo, M., White, P. J., Ding, Z., Kwok, R., Lee, P., Radda, G. K., Toh, H. C., Hirschey, M. D., & Han, W. (2019). Loss of BCAA catabolism during carcinogenesis enhances mTORC1 activity and promotes tumor development and progression. Cell Metabolism, 29(5), 1151–1165.e1156. https://doi.org/10.1016/j.cmet.2018.12.020
Wu, C., Dai, C., Li, X., Sun, M., Chu, H., Xuan, Q., Yin, Y., Fang, C., Yang, F., Jiang, Z., Lv, Q., He, K., Qu, Y., Zhao, B., Cai, K., Zhang, S., Sun, R., Xu, G., Zhang, L., et al. (2022). AKR1C3-dependent lipid droplet formation confers hepatocellular carcinoma cell adaptability to targeted therapy. Theranostics, 12(18), 7681–7698. https://doi.org/10.7150/thno.74974
Park, S., Mossmann, D., Chen, Q., Wang, X., Dazert, E., Colombi, M., Schmidt, A., Ryback, B., Ng, C. K. Y., Terracciano, L. M., Heim, M. H., & Hall, M. N. (2022). Transcription factors TEAD2 and E2A globally repress acetyl-CoA synthesis to promote tumorigenesis. Molecular Cell, 82(22), 4246–4261.e4211. https://doi.org/10.1016/j.molcel.2022.10.027
Funding
Open access funding provided by University of Innsbruck and Medical University of Innsbruck. This work was funded in part by the Austrian Science Fund (FWF) (P 36299, I 4968), the German Research Council (GRK 1715), and the Phospholipid Research Center (AKO-2O22-100/2-2).
Author information
Authors and Affiliations
Contributions
Conceptualization: FS and AK; writing original draft: FS and AK; writing, review, and editing: FS and AK; funding acquisition: AK.
Corresponding author
Ethics declarations
Competing interests
A.K. performs contract research for Bionorica SE, which has a silibinin-containing phytopharmaceutical in their portfolio.
Disclaimer
For the purpose of open access, the authors have applied a CC BY public copyright licence to any author accepted manuscript version arising from this submission.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Su, F., Koeberle, A. Regulation and targeting of SREBP-1 in hepatocellular carcinoma. Cancer Metastasis Rev 43, 673–708 (2024). https://doi.org/10.1007/s10555-023-10156-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-023-10156-5