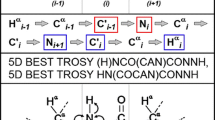
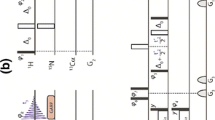
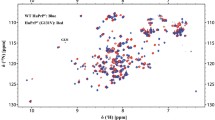
Abstract
Intrinsically disordered proteins (IDPs) are abundant in nature and characterization of their potential structural propensities remains a widely pursued but challenging task. Analysis of NMR secondary chemical shifts plays an important role in such studies, but the output of such analyses depends on the accuracy of reference random coil chemical shifts. Although uniform perdeuteration of IDPs can dramatically increase spectral resolution, a feature particularly important for the poorly dispersed IDP spectra, the impact of deuterium isotope shifts on random coil values has not yet been fully characterized. Very precise 2H isotope shift measurements for 13Cα, 13Cβ, 13C′, 15N, and 1HN have been obtained by using a mixed sample of protonated and uniformly perdeuterated α-synuclein, a protein with chemical shifts exceptionally close to random coil values. Decomposition of these isotope shifts into one-bond, two-bond and three-bond effects as well as intra- and sequential residue contributions shows that such an analysis, which ignores conformational dependence, is meaningful but does not fully describe the total isotope shift to within the precision of the measurements. Random coil 2H isotope shifts provide an important starting point for analysis of such shifts in structural terms in folded proteins, where they are known to depend strongly on local geometry.



Similar content being viewed by others
References
Abildgaard J, Hansen PE, Manalo MN, LiWang A (2009) Deuterium isotope effects on N-15 backbone chemical shifts in proteins. J Biomol NMR 44:119–126
Bertoncini CW, Jung YS, Fernandez CO, Hoyer W, Griesinger C, Jovin TM, Zweckstetter M (2005) Release of long-range tertiary interactions potentiates aggregation of natively unstructured alpha-synuclein. Proc Natl Acad Sci USA 102:1430–1435
Bodner CR, Dobson CM, Bax A (2009) Multiple tight phospholipid-binding modes of alpha-synuclein revealed by solution NMR spectroscopy. J Mol Biol 390:775–790
Camilloni C, De Simone A, Vranken WF, Vendruscolo M (2012) Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry 51:2224–2231
Cavalli A, Salvatella X, Dobson CM, Vendruscolo M (2007) Protein structure determination from NMR chemical shifts. Proc Natl Acad Sci USA 104:9615–9620
Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13:289–302
De Simone A, Cavalli A, Hsu STD, Vranken W, Vendruscolo M (2009) Accurate random coil chemical shifts from an analysis of loop regions in native states of proteins. J Am Chem Soc 131:16332–16333
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRpipe - a multidimensional spectral processing system based on Unix pipes. J Biomol NMR 6:277–293
Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6:197–208
Eghbalnia HR, Wang LY, Bahrami A, Assadi A, Markley JL (2005) Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J Biomol NMR 32:71–81
Eliezer D, Kutluay E, Bussell R, Browne G (2001) Conformational properties of alpha-synuclein in its free and lipid-associated states. J Mol Biol 307:1061–1073
Gardner KH, Rosen MK, Kay LE (1997) Global folds of highly deuterated, methyl-protonated proteins by multidimensional NMR. Biochemistry 36:1389–1401
Garrett DS, Seok YJ, Liao DI, Peterkofsky A, Gronenborn AM, Clore GM (1997) Solution structure of the 30 kDa N-terminal domain of enzyme I of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system by multidimensional NMR. Biochemistry 36:2517–2530
Goddard TD, Kneller DG (2008) Sparky 3. University of California, San Francisco
Griesinger C, Sørensen OW, Ernst RR (1986) Correlation of connected transitions by two-dimensional NMR spectroscopy. J Chem Phys 85:6837–6852
Gronenborn AM, Clore GM (1994) Identification of N-terminal helix capping boxes by means of 13C chemical shifts. J Biomol NMR 4:455–458
Hyberts SG, Milbradt AG, Wagner AB, Arthanari H, Wagner G (2012) Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional poisson gap scheduling. J Biomol NMR 52:315–327
Jameson CJ (1996) Isotope effects on chemical shifts and coupling constants. In: Grant DM, Harris RK (eds) Encyclopedia of nuclear magnetic resonance, vol 4. Wiley, New York, pp 2638–2655
Kazimierczuk K, Stanek J, Zawadzka-Kazimierczuk A, Kozminski W (2010) Random sampling in multidimensional NMR spectroscopy. Prog Nucl Magn Reson Spectrosc 57:420–434
Kjaergaard M, Brander S, Poulsen FM (2011) Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH. J Biomol NMR 49:139–149
Lemaster DM (1994) Isotope labeling in solution protein assignment and structural analysis. Prog Nucl Magn Reson Spectrosc 26:371–419
Lemaster DM, Laiuppa JC, Kushlan DM (1994) Differential deuterium isotope shifts and one-bond 1H–13C scalar couplings in the conformational analysis of protein glycine residues. J Biomol NMR 4:863–870
Maltsev AS, Grishaev A, Bax A (2012a) Monomeric alpha-synuclein binds congo red micelles in a disordered manner. Biochemistry 51:631–642
Maltsev AS, Ying JF, Bax A (2012b) Impact of N-terminal acetylation of α-synuclein on its random coil and lipid binding properties. Biochemistry 51:5004–5013
Marsh JA, Singh VK, Jia ZC, Forman-Kay JD (2006) Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci 15:2795–2804
Neal S, Berjanskii M, Zhang HY, Wishart DS (2006) Accurate prediction of protein torsion angles using chemical shifts and sequence homology. Magn Reson Chem 44:S158–S167
Nietlispach D, Clowes RT, Broadhurst RW, Ito Y, Keeler J, Kelly M, Ashurst J, Oschkinat H, Domaille PJ, Laue ED (1996) An approach to the structure determination of larger proteins using triple resonance NMR experiments in conjunction with random fractional deuteration. J Am Chem Soc 118:407–415
Orekhov VY, Ibraghimov I, Billeter M (2003) Optimizing resolution in multidimensional NMR by three-way decomposition. J Biomol NMR 27:165–173
Ottiger M, Bax A (1997) An empirical correlation between amide deuterium isotope effects on C-13(alpha) chemical shifts and protein backbone conformation. J Am Chem Soc 119:8070–8075
Ozenne V, Bauer F, Salmon L, Huang J-r, Jensen MR, Segard S, Bernado P, Charavay C, Blackledge M (2012) Flexible-meccano: a tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics 28:1463–1470
Peti W, Smith LJ, Redfield C, Schwalbe H (2001) Chemical shifts in denatured proteins: resonance assignments for denatured ubiquitin and comparisons with other denatured proteins. J Biomol NMR 19:153–165
Rezaei-Ghaleh N, Blackledge M, Zweckstetter M (2012) Intrinsically disordered proteins: from sequence and conformational properties toward drug discovery. ChemBioChem 13:930–950
Richarz R, Wuthrich K (1978) 13C NMR chemical shifts of common amino acid residues measured in aqueous solutions of linear tetrapeptides H-GLY-GLY-X-L-ALA-OH. Biopolymers 17:2133–2141
Romero P, Obradovic Z, Li XH, Garner EC, Brown CJ, Dunker AK (2001) Sequence complexity of disordered protein. Proteins Struct Funct Genet 42:38–48
Rovnyak D, Frueh DP, Sastry M, Sun ZYJ, Stern AS, Hoch JC, Wagner G (2004) Accelerated acquisition of high resolution triple-resonance spectra using non-uniform sampling and maximum entropy reconstruction. J Magn Reson 170:15–21
Schwarzinger S, Kroon GJA, Foss TR, Chung J, Wright PE, Dyson HJ (2001) Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc 123:2970–2978
Shen Y, Bax A (2010) Prediction of Xaa-Pro peptide bond conformation from sequence and chemical shifts. J Biomol NMR 46:199–204
Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu GH, Eletsky A, Wu YB, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A (2008) Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA 105:4685–4690
Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS+ : a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44:213–223
Tamiola K, Acar B, Mulder FAA (2010) Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J Am Chem Soc 132:18000–18003
Uversky VN, Dunker AK (2010) Understanding protein non-folding. BBA-Proteins. Proteomics 1804:1231–1264
Venters RA, Farmer BT, Fierke CA, Spicer LD (1996) Characterizing the use of perdeuteration in NMR studies of large proteins C-13, N-15 and H-1 assignments of human carbonic anhydrase II. J Mol Biol 264:1101–1116
Wang YJ, Jardetzky O (2002) Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci 11:852–861
Wang Y, Fisher JC, Mathew R, Ou L, Otieno S, Sublet J, Xiao L, Chen J, Roussel MF, Kriwacki RW (2011) Intrinsic disorder mediates the diverse regulatory functions of the Cdk inhibitor p21. Nat Chem Biol 7:214–221
Wishart DS, Sykes BD (1994) The C-13 chemical shift index—a simple method for the identification of protein secondary structure using C-13 chemical shift data. J Biomol NMR 4:171–180
Wishart DS, Sykes BD, Richards FM (1991) Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol 222:311–333
Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD (1995) 1H, 13C and 15 N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR 5:67–81
Wishart DS, Arndt D, Berjanskii M, Tang P, Zhou J, Lin G (2008) CS23D: a web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res 36:496–502
Ying JF, Chill JH, Louis JM, Bax A (2007) Mixed-time parallel evolution in multiple quantum NMR experiments: sensitivity and resolution enhancement in heteronuclear NMR. J Biomol NMR 37:195–204
Acknowledgments
We thank James L. Baber for experimental support. This work was funded by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (NIH) and the Intramural AIDS-Targeted Antiviral Program of the Office of the Director, NIH.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Maltsev, A.S., Ying, J. & Bax, A. Deuterium isotope shifts for backbone 1H, 15N and 13C nuclei in intrinsically disordered protein α-synuclein. J Biomol NMR 54, 181–191 (2012). https://doi.org/10.1007/s10858-012-9666-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-012-9666-x