Abstract
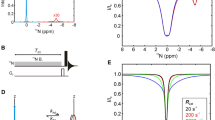
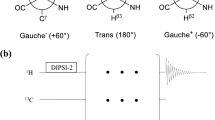
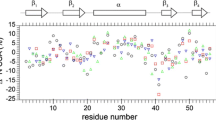
A new pulse sequence is presented for the measurement of relaxation dispersion profiles quantifying millisecond time-scale exchange dynamics of side-chain carbonyl groups in uniformly 13C labeled proteins. The methodology has been tested using the 87-residue colicin E7 immunity protein, Im7, which is known to fold via a partially structured low populated intermediate that interconverts with the folded, ground state on the millisecond time-scale. Comparison of exchange parameters extracted for this folding ‘reaction’ using the present methodology with those obtained from more ‘traditional’ 15N and backbone carbonyl probes establishes the utility of the approach. The extracted excited state side-chain carbonyl chemical shifts indicate that the Asx/Glx side-chains are predominantly unstructured in the Im7 folding intermediate. However, several crucial salt-bridges that exist in the native structure appear to be already formed in the excited state, either in part or in full. This information, in concert with that obtained from existing backbone and side-chain methyl relaxation dispersion experiments, will ultimately facilitate a detailed description of the structure of the Im7 folding intermediate.






Similar content being viewed by others
References
Auer R, Neudecker P, Muhandiram DR, Lundstrom P, Hansen DF, Konrat R, Kay LE (2009) Measuring the signs of 1H(alpha) chemical shift differences between ground and excited protein states by off-resonance spin-lock R(1rho) NMR spectroscopy. J Am Chem Soc 131:10832–10833
Baldwin AJ, Kay LE (2009) NMR spectroscopy brings invisible protein states into focus. Nat Chem Biol 5:808–814
Boehr DD, McElheny D, Dyson HJ, Wright PE (2006) The dynamic energy landscape of dihydrofolate reductase catalysis. Science 313:1638–1642
Bouvignies G, Korzhnev DM, Neudecker P, Hansen DF, Cordes MH, Kay LE (2010) A simple method for measuring signs of (1)H(N) chemical shift differences between ground and excited protein states. J Biomol NMR 47:135–141
Capaldi AP, Shastry MC, Kleanthous C, Roder H, Radford SE (2001) Ultrarapid mixing experiments reveal that Im7 folds via an on-pathway intermediate. Nat Struct Biol 8:68–72
Capaldi AP, Kleanthous C, Radford SE (2002) Im7 folding mechanism: misfolding on a path to the native state. Nat Struct Biol 9:209–216
Cavalli A, Salvatella X, Dobson CM, Vendruscolo M (2007) Protein structure determination from NMR chemical shifts. Proc Natl Acad Sci USA 104:9615–9620
Chak KF, Safo MK, Ku WY, Hsieh SY, Yuan HS (1996) The crystal structure of the immunity protein of colicin E7 suggests a possible colicin-interacting surface. Proc Natl Acad Sci USA 93:6437–6442
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Dennis CA, Videler H, Pauptit RA, Wallis R, James R, Moore GR, Kleanthous C (1998) A structural comparison of the colicin immunity proteins Im7 and Im9 gives new insights into the molecular determinants of immunity-protein specificity. Biochem J 333(Pt 1):183–191
Esadze A, Li DW, Wang T, Bruschweiler R, Iwahara J (2011) Dynamics of lysine side-chain amino groups in a protein studied by heteronuclear 1H–15 N NMR spectroscopy. J Am Chem Soc 133:909–919
Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D, Alber T (2009) Hidden alternative structures of proline isomerase essential for catalysis. Nature 462:669–673
Geen H, Freeman R (1991) Band-selective radiofrequency pulses. J Magn Reson 93:93–141
Gerstein M (1997) A structural census of genomes: comparing bacterial, eukaryotic, and archaeal genomes in terms of protein structure. J Mol Biol 274:562–576
Goddard TD, Kneller D SPARKY 3. University of California, San Francisco
Gorski SA, Le Duff CS, Capaldi AP, Kalverda AP, Beddard GS, Moore GR, Radford SE (2004) Equilibrium hydrogen exchange reveals extensive hydrogen bonded secondary structure in the on-pathway intermediate of Im7. J Mol Biol 337:183–193
Hansen DF, Vallurupalli P, Kay LE (2008a) An improved 15 N relaxation dispersion experiment for the measurement of millisecond time-scale dynamics in proteins. J Phys Chem 112:5898–5904
Hansen DF, Vallurupalli P, Kay LE (2008b) Using relaxation dispersion NMR spectroscopy to determine structures of excited, invisible protein states. J Biomol NMR 41:113–120
Hansen DF, Vallurupalli P, Lundstrom P, Neudecker P, Kay LE (2008c) Probing chemical shifts of invisible states of proteins with relaxation dispersion NMR spectroscopy: how well can we do? J Am Chem Soc 130:2667–2675
Hu JS, Bax A (1996) Measurement of three-bond 13C–13C J couplings between carbonyl and carbonyl/carboxyl carbons in isotopically enriched proteins. J Am Chem Soc 118:8170–8171
Ishima R, Torchia DA (1999) Estimating the time scale of chemical exchange of proteins from measurements of transverse relaxation rates in solution. J Biomol NMR 14:369–372
Ishima R, Torchia D (2003) Extending the range of amide proton relaxation dispersion experiments in proteins using a constant-time relaxation-compensated CPMG approach. J Biomol NMR 25:243–248
Ishima R, Baber J, Louis JM, Torchia DA (2004) Carbonyl carbon transverse relaxation dispersion measurements and ms-micros timescale motion in a protein hydrogen bond network. J Biomol NMR 29:187–198
Karplus M, Kuriyan J (2005) Molecular dynamics and protein function. Proc Natl Acad Sci USA 102:6679–6685
Kay LE (1993) Pulsed-field gradient-enhanced three-dimensional NMR experiment for correlating 13Ca/b, 13C’ and 1Ha chemical shifts in uniformly 13C-labeled proteins dissolved in H2O. J Am Chem Soc 115:2055–2057
Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE (2004) Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature 430:586–590
Korzhnev DM, Religa TL, Banachewicz W, Fersht AR, Kay LE (2010) A transient and low-populated protein-folding intermediate at atomic resolution. Science 329:1312–1316
Kupce E, Freeman R (1996) Optimized adiabatic pulses for wideband inversion. J Magn Reson Ser A 118:299–303
Kupce E, Wagner G (1996) Multisite band-selective decoupling in proteins. J Magn Reson B 110:309–312
Le Duff CS, Whittaker SB, Radford SE, Moore GR (2006) Characterisation of the conformational properties of urea-unfolded Im7: implications for the early stages of protein folding. J Mol Biol 364:824–835
Loria JP, Rance M, Palmer AG (1999) A relaxation compensated CPMG sequence for characterizing chemical exchange. J Am Chem Soc 121:2331–2332
Lundstrom P, Kay LE (2009) Measuring 13Cb chemical shifts of invisible excited states in proteins by relaxation dispersion NMR spectroscopy. J Biomol NMR 44:139–155
Lundstrom P, Vallurupalli P, Religa TL, Dahlquist FW, Kay LE (2007) A single-quantum methyl 13C-relaxation dispersion experiment with improved sensitivity. J Biomol NMR 38:79–88
Lundstrom P, Hansen DF, Kay LE (2008) Measurement of carbonyl chemical shifts of excited protein states by relaxation dispersion NMR spectroscopy: comparison between uniformly and selectively (13)C labeled samples. J Biomol NMR 42:35–47
Lundstrom P, Hansen DF, Vallurupalli P, Kay LE (2009) Accurate measurement of alpha proton chemical shifts of excited protein states by relaxation dispersion NMR spectroscopy. J Am Chem Soc 131:1915–1926
Marion D, Ikura M, Tschudin R, Bax A (1989) Rapid recording of 2D NMR spectra without phase cycling. Application to the study of hydrogen exchange in proteins. J Magn Reson 85:393–399
Millet O, Loria JP, Kroenke CD, Pons M, Palmer AG (2000) The static magnetic field dependence of chemical exchange linebroadening defines the NMR chemcial shift time scale. J Am Chem Soc 122:2867–2877
Mulder FA, Akke M (2003) Carbonyl 13C transverse relaxation measurements to sample protein backbone dynamics. Magn Reson Chem 41:853–865
Mulder FAA, Skrynnikov NR, Hon B, Dahlquist FW, Kay LE (2001) Measurement of slow timescale dynamics in protein sidechains by 15 N relaxation dispersion NMR spectroscopy: Application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J Am Chem Soc 123:967–975
Orekhov VY, Korzhnev DM, Kay LE (2004) Double- and zero-quantum NMR relaxation dispersion experiments sampling millisecond time scale dynamics in proteins. J Am Chem Soc 126:1886–1891
Palmer AG, Kroenke CD, Loria JP (2001) NMR methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzym 339:204–238
Press WH, Flannery BP, Teukolsky SA, Vetterling WT (1988) Numerical recipes in C. Cambridge University Press, Cambridge
Shen Y, Bax A (2010) SPARTA + : a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J Biomol NMR 48:13–22
Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS +: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44:213–223
Skrynnikov NR, Mulder FAA, Hon B, Dahlquist FW, Kay LE (2001) Probing slow time scale dynamics at methyl-containing side chains in proteins by relaxation dispersion NMR measurements: Application to methionine residues in a cavity mutant of T4 lysozyme. J Am Chem Soc 123:4556–4566
Skrynnikov NR, Dahlquist FW, Kay LE (2002) Reconstructing NMR spectra of “invisible” excited protein states using HSQC and HMQC experiments. J Am Chem Soc 124:12352–12360
Tollinger M, Skrynnikov NR, Mulder FAA, Forman-Kay JD, Kay LE (2001) Slow dynamics in folded and unfolded states of an SH3 domain. J Am Chem Soc 123:11341–11352
Tollinger M, Kay LE, Forman-Kay JD (2005) Measuring pK(a) values in protein folding transition state ensembles by NMR spectroscopy. J Am Chem Soc 127:8904–8905
Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Wenger RK, Yao H, Markley JL (2008) BioMagResBank. Nucleic Acids Res 36: D402–D408
Vallurupalli P, Hansen DF, Stollar EJ, Meirovitch E, Kay LE (2007) Measurement of bond vector orientations in invisible excited states of proteins. Proc Natl Acad Sci USA 104:18473–18477
Vuister GW, Bax A (1992) Resolution enhancement and spectral editing of uniformly 13C-enriched proteins by homonuclear broadband 13C decoupling. J Magn Reson 98:428–435
Whittaker SB, Spence GR, Grossmann JG, Radford SE, Moore GR (2007) NMR analysis of the conformational properties of the trapped on-pathway folding intermediate of the bacterial immunity protein Im7. J Mol Biol 366:1001–1015
Wishart DS, Arndt D, Berjanskii M, Tang P, Zhou J, Lin G (2008) CS23D: a web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res 36:W496–W502
Acknowledgments
The authors thank Dr. Sara Whittaker and Professors Geoff Moore and Sheena Radford for the gift of Im7 constructs. A.L.H. acknowledges the National Science Foundation for post-doctoral support (OISE-0852964). This work was supported by grants from the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada (LEK). L.E.K. holds a Canada Research Chair in Biochemistry.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10858_2011_9520_MOESM1_ESM.pdf
Supplementary material 1 (PDF 272 kb). Table of side-chain carbonyl \( |\Updelta \varpi | \) values for the Im7 protein along with pulse sequence code for recording side-chain carbonyl CPMG relaxation dispersion profiles in U-[13C] labeled proteins
Rights and permissions
About this article
Cite this article
Hansen, A.L., Kay, L.E. Quantifying millisecond time-scale exchange in proteins by CPMG relaxation dispersion NMR spectroscopy of side-chain carbonyl groups. J Biomol NMR 50, 347–355 (2011). https://doi.org/10.1007/s10858-011-9520-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-011-9520-6