Abstract
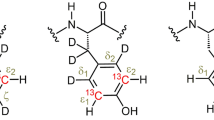
A pulse sequence is described for recording single-quantum 13C-methyl relaxation dispersion profiles of 13C-selectively labeled methyl groups in proteins that offers significant improvements in sensitivity relative to existing approaches where initial magnetization derives from 13C polarization. Sensitivity gains in the new experiment are achieved by making use of polarization from 1H spins and 1H → 13C → 1H type magnetization transfers. Its utility has been established by applications involving three different protein systems ranging in molecular weight from 8 to 28 kDa, produced using a number of different selective labeling approaches. In all cases exchange parameters from both 13C→1H and 1H → 13C → 1H classes of experiment are in good agreement, with gains in sensitivity of between 1.7 and 4-fold realized using the new scheme.







Similar content being viewed by others
References
Allen M, Friedler A, Schon O, Bycroft M (2002) The structure of an FF domain from human HYPA/FBP11. J Mol Biol 323:411–416
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRpipe—a multidimensional spectral processing system based on unix pipes. J Biomol NMR 6:277–293
Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D (2005) Intrinsic dynamics of an enzyme underlines catalysis. Nature 438:117–121
Feher VA, Baldwin EP, Dahlquist FW (1996) Access of ligands to cavities within the core of a protein is rapid. Nat Struct Biol 3:516–521
Geen H, Freeman R (1991) Band-selective radiofrequency pulses. J Magn Reson 93:93–141
Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE (1999) A robust and costeffective method for the production of Val, Leu, Ile (δ1) methyl-protonated N-15-, C-13-, H-2-labeled proteins. J Biomol NMR 13:369–374
Gryk MR, Jardetzky O, Klig LS, Yanofsky C (1996) Flexibility of DNA binding domain of trp repressor required for recognition of different operator sequences. Protein Sci 5:1195–1197
Hill RB, Bracken C, DeGrado WF, Palmer AG (2000) Molecular motions and protein folding: characterization of the backbone dynamics and folding equilibrium of αD-2 using C-13 NMR spin relaxation. J Am Chem Soc 122:11610–11619
Ishima R, Torchia DA (2000) Protein dynamics from NMR. Nat Struct Biol 7:740–743
Ishima R, Torchia DA (2003) Extending the range of amide proton relaxation dispersion experiments in proteins using a constant-time relaxation-compensated CPMG approach. J Biomol NMR 25:243–248
Jemth P, Day R, Gianni S, Khan F, Allen M, Daggett V, Fersht AR (2005) The structure of the major transition state for folding of an FF domain from experiment and simulation. J Mol Biol 350:363–378
Kalodimos CG, Biris N, Bonvin AMJJ, Levandoski MM, Guennuegues M, Boelens R, Kaptein R (2004) Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science 305:386–389
Korzhnev DM, Kloiber K, Kanelis V, Tugarinov V, Kay LE (2004a) Probing slow dynamics in high molecular weight proteins by methyl-TROSY NMR spectroscopy: application to a 723-residue enzyme. J Am Chem Soc 126:3964–3973
Korzhnev DM, Kloiber K, Kay LE (2004b) Multiple-quantum relaxation dispersion NMR spectroscopy probing millisecond time-scale dynamics in proteins: theory and application. J Am Chem Soc 126:7320–7329
Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE (2004c) Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature 430:586–590
Loria JP, Rance M, Palmer AG (1999) A relaxation-compensated Carr–Purcell Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J Am Chem Soc 121:2331–2332
Marion D, Ikura M, Tschudin R, Bax A (1989) Rapid recording of 2D NMR-spectra without phase cycling—application to the study of hydrogen-exchange in proteins. J Magn Reson 85:393–399
Millet O, Loria JP, Kroenke CD, Pons M, Palmer AG (2000) The static magnetic field dependence of chemical exchange linebroadening defines the NMR chemical shift time scale. J Am Chem Soc 122:2867–2877
Mulder FAA, Hon B, Mittermaier A, Dahlquist FW, Kay LE (2002) Slow internal dynamics in proteins: application of NMR relaxation dispersion spectroscopy to methyl groups in a cavity mutant of T4 lysozyme. J Am Chem Soc 124:1443–1451
Mulder FAA, Mittermaier A, Hon B, Dahlquist FW, Kay LE (2001a) Studying excited states of proteins by NMR spectroscopy. Nat Struct Biol 8:932–935
Mulder FAA, Skrynnikov NR, Hon B, Dahlquist FW, Kay LE (2001b) Measurement of slow (μs-ms) time scale dynamics in protein side chains by N-15 relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J Am Chem Soc 123:967–975
Neidhardt FC, Bloch PL, Smith DF (1974) Culture Medium for Enterobacteria. J Bacteriol 119:736–747
Palmer AG, Williams J, McDermott A (1996) Nuclear magnetic resonance studies of biopolymer dynamics. J Phys Chem 100:13293–13310
Piotto M, Saudek V, Sklenar V (1992) Gradient-tailored excitation for single-quantum NMR-spectroscopy of aqueous solutions. J Biomol NMR 2:661–665
Popovych N, Sun SJ, Ebright RH, Kalodimos CG (2006) Dynamically driven protein allostery. Nat Struct Mol Biol 13:831–838
Press WH, Flannery BP, Teukolsky SA, Vetterling WT (1988) Numerical Recipes in C. Cambridge University Press, Cambridge
Shaka AJ, Keeler J, Frenkiel T, Freeman R (1983) An improved sequence for broad band decoupling-WALTZ-16. J Magn Reson 52:335–338
Skrynnikov NR, Mulder FAA, Hon B, Dahlquist FW, Kay LE (2001) Probing slow time scale dynamics at methyl-containing side chains in proteins by relaxation dispersion NMR measurements: application to methionine residues in a cavity mutant of T4 lysozyme. J Am Chem Soc 123:4556–4566
Sprangers R, Kay LE (2007) Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature 445:718–722
Tollinger M, Skrynnikov NR, Mulder FAA, Forman-Kay JD, Kay LE (2001) Slow dynamics in folded and unfolded states of an SH3 domain. J Am Chem Soc 123:11341–11352
Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE (2003) Cross-correlated relaxation enhanced H-1-C-13 NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc 125:10420–10428
Vallurupalli P, Kay LE (2006) Complementarity of ensemble and single-molecule measures of protein motion: a relaxation dispersion NMR study of an enzyme complex. Proc Natl Acad Sci USA 103:11910–11915
Yang H, Luo GB, Karnchanaphanurach P, Louie TM, Rech I, Cova S, Xun LY, Xie XS (2003) Protein conformational dynamics probed by single-molecule electron transfer. Science 302:262–266
Acknowledgments
P. L. and P. V. are supported by fellowships from the Hellmuth Hertz foundation and the Canadian Institutes of Health Research (CIHR) Training Grant in Protein Folding and Disease. This research was supported by a grant from the CIHR. L.E.K. holds a Canada Research Chair in Biochemistry.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lundström, P., Vallurupalli, P., Religa, T.L. et al. A single-quantum methyl 13C-relaxation dispersion experiment with improved sensitivity. J Biomol NMR 38, 79–88 (2007). https://doi.org/10.1007/s10858-007-9149-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-007-9149-7