Abstract
Understanding factors that limit gene flow through the landscape is crucial for conservation of organisms living in fragmented habitats. We analysed patterns of gene flow in Elater ferrugineus, an endangered click beetle living in old-growth, hollow trees in a network of rural avenues surrounded by inhospitable arable land. Using amplified fragment length polymorphism (AFLP) data, we aimed to evaluate if the landscape features important for the beetle’s development are also important for its dispersal. By dividing the sampling area into 30 × 30 m cells, with each cell categorised into one of four classes according to its putative permeability for dispersing beetles, and by correlating matrices of genetic and landscape resistance distances, we evaluated which of the landscape models had the best fit with the observed kinship structure. Significant correlations between genetic and Euclidean distances were detected, which indicated that restricted dispersal is the main constraint driving differentiation between populations of E. ferrugineus. Out of 81 landscape models in total, 54 models yielded significantly weaker correlation between matrices of pairwise kinship and effective distances than the null model. Regression analysis pointed to avenues as having the highest and positive impact on the concordance between matrices of kinship and landscape distances, while open arable land had the opposite effect. Our study thus shows that tree avenues can function as efficient dispersal corridors for E. ferrugineus, highlighting the importance of saving such avenues to increase the connectivity among suitable habitat patches, thereby reducing the risk of local extinctions of E. ferrugineus as well as other saproxylic organisms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Habitat fragmentation, with its often detrimental genetic consequences, is considered one of the greatest threats to biodiversity (Fahrig 2003). The task of maintaining biodiversity is especially challenging in modern agricultural landscapes, where most of the available space has been occupied for cultivation of crops and infrastructure related to this activity (Batáry et al. 2011). Only a minor surface in such landscapes (mainly on field borders and road verges) is not converted into arable land and can support flora and fauna (Dover et al. 2000; Ouin and Burel 2002; Walker et al. 2006; Öckinger and Smith 2006; Skórka et al. 2013). Unfortunately, the probability of local extinction increases as patch area decreases and the probability of colonization decreases as patch connectivity decreases (Moilanen and Hanski 1998). Therefore, the main challenges facing conservation biology in agricultural areas are to maintain the quality of existing habitat patches and to increase the connectivity between patches through the creation of ecological corridors (Barrett and Peles 1994). Designing effective systems of ecological corridors, however, requires an understanding about how the landscape structure affects the movement of individuals of different species (Dover and Settele 2009; Spear et al. 2010). Habitat corridors are generally believed to increase dispersal, i.e. the exchange of individuals (and genes) between isolated demes (Tewksbury et al. 2002). Nonetheless, empirical data and methods to analyse which structural characteristics of the agricultural landscape could affect its connectivity are needed for different organisms. Evaluation of landscape features important for dispersal in a particular animal species can be achieved by examination of how the landscape configuration translates into movement of individuals. Since it is difficult to measure dispersal directly at large spatial scales, genetic data are often used instead (Lowe and Allendorf 2010). It should be noted however that genetic methods only allow the measurement of effective dispersal, i.e. those dispersal events that contribute to gene flow (Guerriero et al. 2011). The influence of landscape structure on effective dispersal could be determined by developing resistance surfaces—spatial models, in which specific landscape features are translated into likely costs of dispersal for the organism under study (Spear et al. 2010; Zeller et al. 2012; Baguette et al. 2013). Habitat models can be used to calculate the among-individuals distances corrected for dispersal costs (the so-called ‘effective distances’), and tested by correlating calculated distances with genetic similarity between individuals (Spear et al. 2010).
For measurement of effective distances, a variety of methods have been proposed (see Kool et al. 2012 for a review). Straight-line distance, the simplest measure of the cost of movement between two individuals or sampling sites, does not consider how landscape constraints affect the movement of an organism. Since elements in a landscape mosaic may differ in their resistance to movement, measures taking into account diversified viscosity of the different landscape components seem to be superior. Among them, the isolation-by-resistance approach, based on the theory of electrical circuits and graph theory (Chandra et al. 1996; McRae 2006), is gaining increasing popularity. This approach takes into account the effect of all alternative pathways between habitat patches, which is especially appropriate when the specific aim is to assess gene flow across a landscape over many generations. Several studies indicated that resistance distances may allow for better assessment of genetic isolation-by-distance than the usual Euclidean or least-cost-path distance because the metric is based on assumptions about the impact of many alternative routes in the entire landscape on the dispersal of individuals, and consequently on the chances of gene flow (McRae and Beier 2007; Schwartz et al. 2009; Lozier et al. 2013). Despite the theoretical superiority of isolation-by-resistance approach over distance measures assuming a single optimal path, the performance of the these approaches have rarely been evaluated with empirical data sets at finer spatial scales (Row et al. 2010).
In this work, we aimed to assess the impact of habitat continuity on dispersal and gene flow of Elater ferrugineus—an endangered beetle associated with old-growth, hollow trees, using Amplified Fragment Polymorphism (AFLP, Vos et al. 1995) data. In agricultural landscapes of Europe, the species often inhabits avenues (i.e. rows of trees planted along roads) and other tree habitats (village parks, wood pastures or remnant forest patches), being dependent mainly on larger trees (Musa et al. 2013). Therefore, E. ferrugineus could serve as an umbrella species for protection of old-growth trees and their associated fauna of saproxylic organsims (Andersson et al. 2014). As yet it is not known whether this species, being extremely dependent on old-growth trees, is able to disperse through treeless areas. The study was designed to cope with this question by assessing genetic relatedness of beetles and comparing it with effective distances in the landscape, i.e. the distances corrected in terms of costs resulting from the movement between habitat patches in the landscape. We expected that the observed gene flow at the landscape level could be better explained by landscape resistance distances than simple Euclidian distances.
Up to now little is known about the population genetics of E. ferrugineus, partly due to the cryptic nature of this species. We hypothesised that, given habitat fragmentation at the landscape level, both genetic drift and restricted dispersal can take place in the study species, causing, in turn, population substructuring. We expect that our results will shed light on the dispersal and population biology of E. ferrugineus and provide effective conservation strategies for this threatened beetle.
Materials and methods
Studied species
The red click beetle E. ferrugineus Linnaeus, 1758, is one of the largest of the Central European click beetles (family Elateridae). It develops in the hollows of old, deciduous trees, which are inhabited by larvae of scarab beetles, such as species of the genera Osmoderma, Protaetia and Gnorimus (Burakowski et al. 1985). Larval E. ferrugineus are facultatively carnivorous. It is believed that their main prey are larvae of the large scarabs listed above, given that adult E. ferrugineus are attracted by the sex pheromone of Osmoderma (kairomone effect, Svensson et al. 2004; Larsson and Svensson 2009). However, larvae can also feed on wood mould alone (Tolasch et al. 2007). As a result of massive habitat loss during the last centuries, E. ferrugineus is today considered a rare and vulnerable species throughout its distribution (Nieto and Alexander 2010), which ranges from Spain to the Caucasus and from Italy to the south of Sweden. In Poland the red click beetle is protected by national law and placed in the red book of animals and on the red list of endangered species as vulnerable (Pawłowski et al. 2002). Before the pheromone lures became available for monitoring (Tolasch et al. 2007; Svensson and Larsson 2008; Svensson et al. 2011) it was considered extremely rare. Much of the available data on the occurrence of E. ferrugineus in Poland concerns the nineteenth and first half of the twentieth century (Burakowski et al. 1985). Over the last 50 years this species has been recorded from only about 25 localities (Buchholz and Ossowska 2004). Recently, rural avenues were recognized as the most important habitat for this highly protected species in Poland (Oleksa et al. unpubl.) which may be valid also for other European countries. Due to the ease of pheromone trapping (Kadej et al. 2015), E. ferrugineus has the potential to become a convenient umbrella species for the rich community of saproxylic species, i.e. organisms dependent on dead wood habitats (Andersson et al. 2014).
Area under study
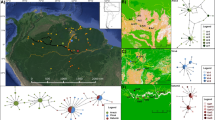
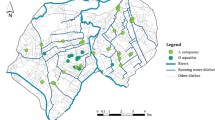
The study was conducted in the municipality of Rychliki in northern Poland (Warmia and Masuria Province, N53° 59′ E19° 31′, Figure 1). Due to fertile soils, the landscape of the area under study is dominated by farmland, occupying 74 % of the surface. Forests cover only 17 % of the area, which is significantly lower than the Polish national average (29 %). Woodlands are retained mainly in gorges and swamps, which are difficult to cultivate. Forests are usually composed of deciduous species, mainly beech Fagus sylvatica, oak Quercus robur, hornbeam Carpinus betulus and—on wet soils—alder Alnus glutinosa. In the studied area, due to intensive management, most forest trees are too young to be a suitable habitat for E. ferrugineus. Despite the high share of farmland in the general area of the municipality and the young age of the forests, the organisms associated with old trees have numerous suitable habitats in the form of roadside avenues of trees and village parks. The avenue network was established in the form similar to the present day in the eighteenth and nineteenth centuries, as shown by maps from that period (von Schrötter 1812). After a habitat inventory in the summer 2011 (Oleksa A., unpublished), we recognized ca. 70 km of rural avenues as suitable habitat for E. ferrugineus and its main prey Osmoderma barnabita. We also found ca. 1500 hollow and thick (DBH ≥ 60 cm) trees potentially inhabited by both species.
Sampling of beetles for molecular analyses
For DNA sampling, beetles were caught with pheromone traps (Tolasch et al. 2007) in the summer 2011. Since the pheromone is attractive only for males, only beetles of this sex were included in the study. Self-made traps baited with the pheromone (Figure 2) were fixed to branches of trees at 3–5 m above ground. Due to the very fast response of males to pheromone exposure and to avoid attracting beetles from long distances, traps were exposed for no more than 1.5 h at a site. At one time, a series of 20 traps were set along a road at 250–500 m intervals (depending on tree density). After checking and emptying the traps, they were moved to the next road section. All catches were conducted during the day (between 11:00 and 18:00), in fine weather (air temperature >20 °C, no rainfall). Sampling covered the entire territory of the municipality of Rychliki, where beetles were trapped in places with abundant mature trees, i.e. along tree-lined roads (avenues). A simulation study by Oyler-McCance et al. (2013) found that linear and systematic sampling regimes performed well in order to detect the effects of landscape pattern on gene flow.
A pheromone-baited funnel trap used for collecting Elater ferrugineus; a two black PCV sheets (25 × 20 cm), properly cut and connected crosswise; b plastic funnel (diameter 25 cm); c container for holding captured insects until their release. Arrow indicate the placement of a 200 µL PCR-tube baited with 2 µL 7-methyloctyl (Z)-4-decenoate, the female-produced sex pheromone of E. ferrugineus
For collection of beetle tissue, the tarsus of one middle leg was cut off with scissors and stored in absolute ethanol. After amputation of tarsi, the beetles were released on the vegetation close to the pheromone trap they were captured in. Altogether, samples from 287 beetles were collected and stored at −20 °C until DNA extraction.
DNA extraction and AFLP genotyping
Genomic DNA was extracted using the Insect Easy DNA Kit (EZNA) (Omega Bio-Tek, Norcross, GA, USA) following the manufacturer’s protocol. The AFLP analysis followed the original protocol by Vos et al. (1995). Restriction-ligation reactions were carried out in a total volume of 10 μl. A single reaction contained 500 ng of genomic DNA, 5 U EcoRI (MBI Fermentas, Vilnius, Lithuania) and 5 U TruI (MseI isoschizomer) (Fermentas), 1.5 U T4 DNA Ligase (Fermentas), 1× T4 DNA Ligase buffer (Fermentas), 0.05 % BSA, 50 mM NaCl, 0.5 pmol/μl E-Adaptor and 5 pmol/μl M-Adaptor. Reactions were carried out at room temperature overnight and then diluted 5× with H2O in order to obtain PCR matrices (pre-matrix DNA) for pre-selective amplification. Pre-selective amplifications were carried out in 10 μl total volume. A pre-selective PCR mixture contained 2 μl pre-matrix DNA, 1× Qiagen Master Mix (Qiagen Taq PCR Master Mix Kit; Qiagen, Hilden, Germany), 0.5 μM E-primer (E + A) and 0.5 μM M-primer (M + C). Amplification was carried out using the following program: 72 °C for 2 min, 20 cycles of 94 °C for 20 s, 56 °C for 30 s and 72 °C for 2 min, and finally 60 °C for 30 min. A product of pre-selective PCR was diluted 20 times in order to obtain a PCR matrix for selective amplification (sel-matrix DNA).
Selective amplification was carried out in 10 μl total volumes, consisting of 3 μl sel-matrix DNA, 1× Qiagen Master Mix, 0.5 μM FAM-labelled E-primer (E + ACA or E + ACG) and 0.5 μM M-primer (M + CAC or M + CAG). Three combinations of primers were used: E_ACA × M_CAC, E_ACA × M_CAG, E_ACG × M_CAC. PCR reaction was performed with the following program: 94 °C for 2 min, 10 cycles of 94 °C for 20 s, 66 °C (−1° per cycle) for 30 s and 72 °C for 2 min, 20 cycles of 94 °C for 30 s, 56 °C for 30 s and 60 °C for 30 min. Both pre-selective and selective amplifications were carried out using a PTC200 thermocycler (BioRad, Hercules, CA, USA).
The products of selective amplifications were sized using an ABI3130XL automated capillary sequencer (Applied Biosystems, Foster City, CA, USA) and the manufacturer’s Genescan 3.7 software. AFLP profiles were then subjected to visual assessment in order to eliminate outlier samples using the Genotyper 3.7 software (Applied Biosystems). Then, all peaks in a profile within the range 60–400 bp were automatically labelled and bins were created based on all labelled peaks. Automatically created bins were visually checked to ensure that the bin was centred on the distribution of peaks within the bin. Bins with low polymorphism (5 % <frequency of dominant haplotype <95 %) were omitted from further study. To reduce the occurrence of homoplasy (Vekemans et al. 2002) bins with fragment-length distributions that overlapped with adjacent bins were also removed. Raw peak intensity data output from Genotyper was subsequently transformed into a binary data matrix with AFLPScore R-script (Whitlock et al. 2008). Given that our main objective was to investigate spatial genetic structure and it may arise both due to isolation-by-distance or due to spatial variation of selective forces affecting the distribution of alleles of specific loci (Beaumont and Balding 2004), only neutral loci were selected for the analyses. For this purpose, we used Mcheza (Antao and Beaumont 2011) to identify outlier loci in terms of their heterozygosity versus F ST , i.e. the loci considered to be candidates for directional or stabilizing selection. Locus-specific F ST values were estimated from a simulated distribution of 50,000 iterations using an infinite alleles model. The resulting distribution was used to find outliers. Outlier loci falling above the 0.95 quantile were considered putatively under directional selection, while under stabilizing selection when falling under 0.05. These markers were omitted from further analyses.
Genetic variation
In the studied landscape, beetles are distributed more or less continuously, mainly in avenues along roads (Figure 1). However, for the assessment of genetic variation we assumed that the beetles isolated by more than 1 km from other sampling sites formed a distinct group of individuals (deme). To avoid bias resulting from small sample size, for estimation of variation we included only sites where at least five individuals were sampled (Figure 1; Table 1). For each deme, after estimating allelic frequencies with a Bayesian method, assuming a non-uniform prior distribution of allele frequencies (AFLP-SURV ver.1.0, Vekemans et al. 2002), the following statistics were computed: number and proportion of polymorphic loci at the 5 % level; expected heterozygosity or Nei’s gene diversity (Hj) and Wright’s F ST .
Relationship between genetic and Euclidean distances
The relationship between pairwise genetic and geographic distances could be examined at two levels, i.e. for populations and for individuals (Manel et al. 2003). The first approach assumes a discrete population subdivision, while the analysis based on individuals is free of assumptions about the spatial scale of structuring in populations and could be more adequate if individuals are distributed continuously across the landscape and/or the borders between populations are unknown, as it was in the studied system.
The relationship between matrices of Euclidean distances and genetic similarities was tested by Mantel correlation with 10,000 permutations (computed with ecodist version 1.2.9 in R version 3.1.0, Goslee and Urban 2007). Additionally, we applied analysis of spatial genetic structure (SGS) based on pairwise kinship coefficients (Hardy 2003) between individuals using the software SPAGeDi ver. 1.3 (Hardy and Vekemans 2002). To visualize the strength of SGS, average pair-wise kinship coefficients (F ij ) were plotted against distance classes (d ij ). The number of distance classes was determined following Sturge’s rule (Sturges 1926) based on the number of all pairwise comparisons and the mean distance per distance class was used to draw the plot. Confidence intervals around observed average kinship coefficients for a given distance class were obtained from standard errors by jackknifing data over loci (20,000 permutations).
Under isolation-by-distance, given drift-dispersal equilibrium, kinship is a linear function of the logarithm of distance between individuals (Rousset 2000). Therefore, in order to illustrate the intensity of the spatial genetic structure, we used the formula S p = −b 1/(1 − f (1)) (Vekemans and Hardy 2004) where b1 is a slope of a log-linear regression between observed kinship and a distance between individuals, and f (1) is the average kinship for the first distance class. Given that Sp is inversely proportional to the effective size of the Wright’s neighbourhood (N b ), the latter parameter was also calculated. The confidence intervals were estimated using the standard error calculated by jackknifing over loci. Finally, the square root of the axial variance of dispersal (σ) was approximated using an iterative procedure implemented in SPAGeDi, based on the regression restricted to a distance range from σ to 20σ (Rousset 1997). As the effective density (D e , expressed as number of individuals per area) remains unknown, a series of guesses on D e were used in the estimation.
Relationship between genetic and landscape (resistance) distances
To evaluate if the landscape features important for the beetle’s development are also used for its dispersal, we developed spatial (raster) models, in which landscape features were translated into likely costs of dispersal for the red click beetle. The area used for the analyses was the minimum convex polygon enclosing all of the sample points, with a 2 km buffer surrounding this polygon added to minimize the increase of resistance values due to the grid boundary (Koen et al. 2010). We assigned a ‘no data’ value to cells outside of this area and excluded them from all calculations.
Our raster models were based on freely available Landsat data with the resolution of 30 m (http://landsat.usgs.gov, years 2010–2012). Although the resolution of the adapted grid can obviously affect the estimation of effective landscape distances, we believe that the adopted grid resolution was appropriate because it is much smaller than the putative dispersal distances of the study organism, as recommended by Anderson et al. (2010). Because E. ferrugineus is considered to be a relatively good flier, movements of several hundred meters are highly possible in this species.
Raster cells were classified according to their putative suitability for E. ferrugineus. We distinguished four grid cell classes, differing in the quantity of old growth-trees: (1) cells containing avenue stretches (hereafter, called “avenues”), (2) cells containing forest patches (“forest”), (3) cells containing a landscape mosaic with thickets or smaller trees but without avenues or forest (“thicket”), and (4) land without any trees (usually arable fields, meadows and water bodies; “open land”). Cells were classified as “avenues” if any fragment of an avenue was placed within the cell; other cells were classified according to the dominant land cover type. Classification of raster cells was performed in two steps. First, using the SAGA software we classified Landsat images into two classes: (1) containing tree cover (forest, thickets or linear plantings of trees), (2) open areas. Next, raster was converted into shape format and classification of polygons was verified in Quantum GIS (QGIS Development Team 2012) based on more detailed aerial images from Google Earth and geoportal.gov.pl. In this step, areas with woody vegetation cover were categorised as forests, avenues and areas that do not fit into any of these classes (“thicket”). The categorization was done independently by two authors (A.O. and R.G.), then—in doubtful cases—its accuracy was examined directly in the field.
In the next step, we elaborated a number of models in which these four cell classes were assigned values describing their permeability for dispersing beetles (the so-called cell conductances, McRae 2006). To minimize subjectivity in assigning conductance values, we assumed that each class could have one of three conductance values (weak—1, moderate—50, or high—100) and created models with all possible combinations of the three conductance values for the four cell classes. As a result we obtained 81 (i.e. 34) landscape grid models, describing all possible conductance surfaces for beetles moving through the landscape (Supplementary data). Subsequently, we computed matrices of pairwise distances between local populations or individuals for each landscape conductance surface, using resistance distance, as implemented in the R package ‘gdistance’ ver. 1.1-5 (van Etten 2014) in the R version 3.1.0 (R Development Core Team 2013). The resistance distance is computed as the random-walk commute time between points (Chandra et al. 1996; McRae 2006). During calculations, we used a connection scheme where movement was allowed between the nearest 16 cells. This scheme is the most realistic scenario, because it assumes that the narrow linear structures with a width of one cell (for example, avenues) do not constitute a strong barrier for movement if their conductance is set to low value.
Because the probability of moving through the cell system is not dependent on the absolute cell values but on the relative differences between them, three combinations of conductance values (when conductance values for all four cell types were set equally to 1, 50 or 100), gave the same model representing the homogeneous landscape, i.e. the analogue of the pure isolation-by-distance model (Wright 1943), which could be regarded as the null model.
To evaluate which of the proposed landscape models had the best fit with the observed pattern of gene flow, we calculated Mantel correlation between the matrix of pairwise genetic similarity coefficients and matrices of effective landscape distances. The genetic similarity matrix was computed using kinship coefficients between pairs of individuals (computed with SPAGeDi version 1.4, Hardy and Vekemans 2002), because the individual level better corresponds to the resolution of the landscape rasters. On the other hand, using discrete demes as data units would lead to data loss and ambiguity, e.g. in determining distances between demes. Mantel tests with 10,000 permutations were calculated using the package ‘ecodist’ version 1.2.7 (Goslee and Urban) in the R version 3.1.0 (R Development Core Team 2013).
To get insight into which land use categories have the most influence on the observed pattern of isolation-by-distance, we performed multiple linear regression with the square of the estimated Mantel correlation coefficient (r 2 M ) as a response variable and conductances of the four landscape types as explanatory variables. We considered all possible (i.e. 15) candidate models that emerged from all combinations of explanatory variables, and these were ranked according to the relative strength of support for each model using Akaike’s information criterion adjusted for small sample sizes (AICc). We used AICc weights (ωi) to generate weighted model-averaged parameter estimates. We also estimated the relative importance of the predictor variables by summing the AICc weights over all the models in which the variable was contained. Parameter estimates and AICc for all models were calculated using the package ‘MuMIn’ version 1.10.0 (Bartoń 2013) in the R version 3.1.0 (R Development Core Team 2013).
Results
Marker polymorphism and genetic variation
Successful amplification was obtained in 247 samples of E. ferrugineus. Three combinations of primers yielded amplification of 297 putative loci (peaks visible in the electropherogram). From this number, 181 were omitted from further analyses either because of the difficulty to distinguish between two or more fragments of a similar mass or due to a vague amplification. For further examination we used 116 loci (E_ACA × M_CAC—35, E_ACA × M_CAG—27 and E_ACG × M_CAC—54 loci), but the F ST -outlier method revealed that 11 loci were positively exceeding neutral expectations, leaving 105 putatively neutral loci for final analyses (E_ACA × M_CAC—34, E_ACA × M_CAG—27 and E_ACG × M_CAC—44 loci).
Demes distinguished with the 1 km-distance criterion showed similar levels of expected heterozygosities, ranging from 0.254 to 0.334 (Table 1). On average, gene diversity within populations amounted to 0.294 (SE 0.009). The proportion of polymorphic loci ranged from 72.4 to 97.1 %. The average pairwise F ST across all populations was equal to 0.067 and ranged from 0 to 0.133.
Relationship between genetic and Euclidean distances
Mantel tests showed that matrices of kinship and Euclidean distances between sampled individuals were significantly correlated [r M = −0.263, 95 % CL (−0.281; −0.246), p < 0.0001]. Similarly, the analysis of autocorrelation of the kinship coefficient in SpaGeDi revealed a significant spatial genetic structure (Figure 3). All descriptive measures of spatial structure, i.e. the kinship among individuals collected at the closest distance (f ij(1) = 0.068 ± 0.008), the slope of the log-linear regression (b 1 = −0.030 ± 0.004), and the spatial genetic structure intensity measure (Sp = 0.033 ± 0.01), were significantly different from zero. The nearest neighbours exhibited the highest kinship, which then decreased with increased distance. Up to 3.6 km distance the genotypes were more related than could be expected from their random distribution, then up to 9.1 km distance the pairwise kinship coefficients were in general undistinguishable from zero and over this distance the genotypes were less related than could be expected from the null hypothesis.
Average kinship coefficients between pairs of individuals of Elater ferrugineus plotted against the straight-line distance. The observed value of pairwise kinship coefficient for mean value of each distance class, its 95 % confidence interval obtained under the null hypothesis that genotypes are randomly distributed (shaded area), and standard error obtained by jack-knifing over loci (error bar) are shown. Distance classes (16) were defined using Sturge’s rule so as to include approximately equal number of pairwise comparisons
Estimation of gene flow
Using the iterative procedure implemented in the SPAGeDi software, under the assumption of isolation-by-distance, we estimated the two characteristics of gene flow intensity: the Wright’s neighbourhood size (N b ) and the axial variance of dispersal (σ), the latter being a measure similar to the mean dispersal distance. The results from the analysis are shown in Table 2. Generally, the presence of convergence for the most realistic values of assumed effective density (De) suggests a good agreement between the observed correlograms and the theory of isolation-by-distance in E. ferrugineus. The axial variance of dispersal in the species was estimated to 110–651 m and neighbourhood size to 26–76 individuals for the moderate values of effective density (0.1–10 individual * ha−1).
Relationship between genetic and landscape distances
Mantel correlation between resistance distances between pairs of individuals of E. ferrugineus and their kinship coefficients ranged between −0.031 and −0.254 (all correlation coefficients significant at p < 0.0001, Figure 4). Resistance distances measured over null landscape models (i.e. homogenous landscapes, in which isolation is only due to distance) yielded a correlation coefficient of −0.247 (95 % CL from −0.264 to −0.233). There were six landscape models in which resistance distances showed slightly stronger correlation with kinship than in these null models (r M between −0.249 and −0.254). However, taking into account 95 % confidence limits (from −0.271 to −0.234), none of them were significantly better than the homogenous (null) model.
Results from Mantel test showing correlations between matrix of pairwise kinship coefficients and matrices of landscape resistance distances, computed for 81 combinations of the three assumed conductance values (either 1, 50 or 100) for the four landscape features. Bars indicate 95 % confidence intervals
On the other hand, 54 raster models showed significantly weaker correlation than the null homogenous model. When comparing models of lower fit with models better matched to the observed pattern of gene flow, it turned out that models equivalent to the null model were characterized by a higher average conductance of avenues than inferior models (respectively, 83.33 and 36.67; the difference was significant when assessed with the Mann–Whitney test, p < 0.0001). In line with this observation, multiple regression (Tables 3, 4) pointed to avenues as a habitat type that have the highest positive impact on landscape connectivity for E. ferrugineus, i.e. the concordance between matrices of pairwise kinship and resistance landscape distances increased with increasing conductance of avenues (relative variable importance equal to 1, β = 0.684, p < 0.001). A similar but weaker effect was evident for the thickets (relative variable importance 0.999, β = 0.304, p < 0.001). In contrast, open land had the opposite effect (relative variable importance 0.864, β = −0.177, p < 0.05), while the effect of forest was not statistically significant (relative variable importance 0.313, β = −0.062, n.s.).
Discussion
In our study, any assumption about conductivity of landscape features for dispersing E. ferrugineus gave better fit between genetic and landscape distances than the simplest homogenous models, in which isolation results from the distance only. Although six models (all but one assuming the highest conductance of avenues and low to moderate conductance of open field) showed slightly stronger correlation with kinship than homogenoues null models, the difference was not significant. On the other hand, models with the opposite setup (low conductance of avenues and higher conductance of open areas) were significantly less correlated with the observed pattern of kinship between individuals. It indicates that the arrangement and relative proportions of habitat and non-habitat patches significantly affect the phenomenon of gene flow in E. ferrugineus. In fact, for many pairwise comparisons it was impossible to differentiate straight-line gene flow from gene flow through the linear habitat, because both distances were virtually the same. Therefore, for shorter distances (within a single avenue) isolation-by-resistance could be reduced to isolation by the Euclidean distance. This is probably the reason why the models of gene flow through the avenues and along straight lines are equally well correlated with the observed pattern of kinship.
The results presented here are in line with the hypothesis that avenues may constitute important landscape feature supporting gene movement in E. ferrugineus. Recently, plantings of trees along roads were recognized as an important habitat for several vulnerable, tree-dependent insects in cultural landscapes (Oleksa et al. 2006, 2007, 2013b; Horák et al. 2010; Michalcewicz and Ciach 2012). However, it remained unclear whether avenues can be considered exclusively as their habitat or could also serve as effective dispersal corridors. Although we were not able to prove that the pattern of gene flow was better fitted with effective landscape distances than with straight-line distances, our results indicate that avenues alone are sufficient to ensure relatively high levels of gene exchange. Since the models with high conductance of avenues and lower conductance of other landscape features were as good as the models assuming homogenous space in explaining the observed pattern of kinship, we may conclude that a network of avenues is sufficient to maintain relatively high level of gene flow, even if a large proportion of the landscape is composed of areas in which dispersal is costly.
Our regression analysis pointed to avenues as the main landscape feature increasing the concordance between genetic and effective landscape distances, while the effect of open (mostly arable) land was the opposite. These effects are consistent with what can be expected in a species strictly dependent on the old-growth tree habitats. However, the impact of forest and thickets needs further explanation. It could be surprising that forests had no significant effect on gene flow in a hollow-dwelling species. Nonetheless, this may result from the fact that forests in the studied area have a long history of management for timber production, which lowered the average age of trees and caused a drastic reduction in the density of mature hollow trees. In contrast to the forests, timber production has never been the primary function of avenues, which were mainly planted in order to protect roads from winds and winter snow (Pradines 2009). Thus, rural avenues have become the main place where mature trees are abundant. Similarly to avenues, our regression analysis identified thickets as one more type of land use which increased the compliance between matrices of kinship and effective landscape distances. In fact, the landscape features which we defined as “thicket” (i.e. raster cells containing a landscape mosaic with thickets or smaller trees but without avenues or forest) were heterogeneous in terms of their composition and/or origin. Nevertheless, a substantial part of this landscape class has developed as a result of degradation of historical avenues through cutting of trees along minor roads, mainly due to intensification of agriculture in the last decades of the twentieth century. Taking into account that the observed spatial genetic structure is a result of historical process of gene dispersal (Hardy and Vekemans 1999), it is therefore possible that the positive effect of thickets is a reflection of the former gene flow, which took place in the past.
Our conclusion is consistent with several previous studies dealing with the role of linear tree or bush habitats as ecological corridors for diverse groups of organisms, including snails (Arnaud 2003), ground beetles (Jopp and Reuter 2005; Jordán et al. 2007), crickets (Berggren et al. 2002; Eriksson et al. 2013) and birds (Gillies and St Clair 2008). In contrast, some other studies did not provide evidence for a beneficial effect of corridors (Haddad et al. 2003; Öckinger and Smith 2008; Åström and Pärt 2013). Results from these studies demonstrate that the effects of corridors may be influenced by the quality of the adjacent matrix and could differ between organisms and landscapes, depending on conditions to which species or local populations are adapted (Dennis et al. 2013). Usually, the movement between habitats is higher if the organism does not perceive the border as distinct (e.g. between two open habitats) and animals are less likely to move through sharp borders (e.g. between meadow and forest), which could direct the movement along the boundary (Ries and Debinski 2001; Kuefler et al. 2010; Eycott et al. 2012; Bertoncelj and Dolman 2012). Therefore, it may be expected that corridors are more important for organisms living in landscapes composed of contrasting types of environments, which is the case in the studied system, where tree habitats are embedded in the inhospitable matrix and the beetle is strictly associated with a narrow ecological niche.
It could also be anticipated that the presence of corridors should direct populations toward evolution of higher dispersal rates. Theoretical models (Travis and Dytham 1999; Stamps et al. 2005) as well as empirical studies (Merckx et al. 2003; Bonelli et al. 2013) indicate that in highly fragmented landscapes there may be selection against dispersal because dispersing individuals may experience high costs of finding suitable habitat patches. If this is the case for E. ferrugineus in the studied landscape, it is possible that arrangement of suitable habitat as a highly continuous network of tree avenues could increase the exchange of individuals between occupied trees, which in turn should result in less intensive spatial genetic structure and a weaker spatial genetic structure compared to the situation when trees are more scattered in the landscape. Previous studies (Musa et al. 2013) showed that the probability of patch occupancy by E. ferrugineus increased with increasing amount of large trees. Our results indicate that also the spatial configuration of trees in the landscape may contribute to the likelihood of the species’ occurrence. The question whether populations of E. ferrugineus living in more fragmented landscapes show weaker dispersal capacity, stronger spatial genetic structure or more elevated inbreeding levels, opens interesting perspectives for further comparative research. Such a pattern was recently found in the beetles of the genus Osmoderma, a main prey of E. ferrugineus larvae, where differences in dispersal distances between populations from different geographic regions of Europe were detected (Ranius and Hedin 2001; Hedin and Ranius 2002; Dubois et al. 2010; Oleksa et al. 2013a; Chiari et al. 2013; Le Gouar et al. 2014). Since the dispersal in poikilotherms is strictly dependent on temperature, these differences may be the result of climatic factors, as observed in other insects (Cormont et al. 2011; Delattre et al. 2013); however, it is also possible that differences in dispersal capacities could be explained by various selective pressures acting on the beetles in landscapes of different connectivity and habitat size.
In addition to assessing the impact of the landscape on gene flow in E. ferrugineus, we aimed to estimate the extent of gene flow in this species. We have demonstrated that the axial variance of dispersal, a measure very close to the average distance of dispersal, amounted to several hundred meters (110–651 m for moderate values of effective density). Although our estimates are imprecise due to the lack of knowledge about the effective density and (possibly) its variability over the studied area, dispersal of several hundred meters is highly possible in this species. Zauli et al. (2014) found that males of E. ferrugineus moved from the site of first capture covering a median distance of 214 m and approximately 50 % of individuals disperse not further than 250 m. Observed movement distances of other saproxylic beetles could comprise more than 700 m in a species which is believed to be poor flier (Dubois et al. 2010; Oleksa et al. 2013a) and even more than 10 km for species with high dispersal capacities (Jonsson 2003; Williams and Robertson 2008; Drag et al. 2011). In this respect, E. ferrugineus can be regarded as a species with lower dispersal abilities, for which habitat continuity plays an important role.
The data presented here allow us to conclude that avenues should be protected as valuable dispersal corridors for organisms dependent on mature trees. Unfortunately, the current need for traffic safety improvement and infrastructure development has caused large-scale cutting of roadside trees (Pradines 2009), and their decline poses the most serious threat for the whole community of arboreal organisms in farmland areas. We believe that understanding the factors affecting the chances of gene flow in E. ferrugineus would be a step forward in developing effective conservation measures to mitigate the impact of its habitat decline. Taking into account the rapid loss of mature trees in modern European landscapes, we recommend strict protection of existing avenues and planting new ones in places of disturbed continuity.
References
Anderson CD, Epperson BK, Fortin M-J, Holderegger R, James PMA, Rosenberg MS, Scribner KT, Spear S (2010) Considering spatial and temporal scale in landscape-genetic studies of gene flow. Mol Ecol 19:3565–3575. doi:10.1111/j.1365-294X.2010.04757.x
Andersson K, Bergman KO, Andersson F, Hedenström E, Jansson N, Burman J, Winde I, Larsson MC, Milberg P (2014) High-accuracy sampling of saproxylic diversity indicators at regional scales with pheromones: the case of Elater ferrugineus (Coleoptera, Elateridae). Biol Conserv 171:156–166. doi:10.1016/j.biocon.2014.01.007
Antao T, Beaumont MA (2011) Mcheza: a workbench to detect selection using dominant markers. Bioinformatics 27:1717–1718. doi:10.1093/bioinformatics/btr253
Arnaud JF (2003) Metapopulation genetic structure and migration pathways in the land snail Helix aspersa: influence of landscape heterogeneity. Landsc Ecol 18:333–346. doi:10.1023/a:1024409116214
Åström J, Pärt T (2013) Negative and matrix-dependent effects of dispersal corridors in an experimental metacommunity. Ecology 94:72–82. doi:10.1890/11-1795.1
Baguette M, Blanchet S, Legrand D, Stevens VM, Turlure C (2013) Individual dispersal, landscape connectivity and ecological networks. Biol Rev Camb Philos Soc 88:310–326. doi:10.1111/brv.12000
Barrett GGW, Peles JDJ (1994) Optimizing habitat fragmentation: an agrolandscape perspective. Landsc Urban Plan 28:99–105. doi:10.1016/0169-2046(94)90047-7
Bartoń K (2013) MuMIn: Multi-model inference. R package version 1.10.0. Vienna, Austria: R Foundation for Statistical Computing. http://CRAN.R-project.org/package=MuMIn
Batáry P, Báldi A, Kleijn D, Tscharntke T (2011) Landscape-moderated biodiversity effects of agri-environmental management: a meta-analysis. Proc Biol Sci 278:1894–1902. doi:10.1098/rspb.2010.1923
Beaumont MA, Balding DJ (2004) Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol 13:969–980. doi:10.1111/j.1365-294X.2004.02125.x
Berggren A, Birath B, Kindvall O (2002) Effect of corridors and habitat edges on dispersal behavior, movement rates, and movement angles in Roesel’s bush-cricket (Metrioptera roeseli). Conserv Biol 16:1562–1569. doi:10.1046/j.1523-1739.2002.01203.x
Bertoncelj I, Dolman PM (2012) The matrix affects trackway corridor suitability for an arenicolous specialist beetle. J Insect Conserv 17:503–510. doi:10.1007/s10841-012-9533-9
Bonelli S, Vrabec V, Witek M, Barbero F, Patricelli D, Nowicki P (2013) Selection on dispersal in isolated butterfly metapopulations. Popul Ecol 55:469–478. doi:10.1007/s10144-013-0377-2
Buchholz L, Ossowska M (2004) Elater ferrugineus Linnaeus, 1758, Tęgosz rdzawy. In: Głowaciński Z, Nowacki J (eds) Polish red data book of animals, invertebrates. Instytut Ochrony Przyrody PAN, Akademia Rolnicza im. A. Cieszkowskiego, Kraków-Poznań, p 119–120
Burakowski B, Mroczkowski M, Stefańska J (1985) Chrząszcze Coleoptera: Buprestoidea, Elateroidea i Cantharoidea. Katalogi Fauny Pol 23:1–401
Chandra AK, Raghavan P, Ruzzo WL, Smolensky R, Tiwari P (1996) The electrical resistance of a graph captures its commute and cover times. Comput Complex 6:312–340. doi:10.1007/BF01270385
Chiari S, Carpaneto GM, Zauli A, Zirpoli GM, Audisio P, Ranius T (2013) Dispersal patterns of a saproxylic beetle, Osmoderma eremita, in Mediterranean woodlands. Insect Conserv Divers 6:309–318. doi:10.1111/j.1752-4598.2012.00215.x
Cormont A, Malinowska AH, Kostenko O, Radchuk V, Hemerik L, WallisDeVries MF, Verboom J (2011) Effect of local weather on butterfly flight behaviour, movement, and colonization: significance for dispersal under climate change. Biodivers Conserv 20:483–503. doi:10.1007/s10531-010-9960-4
Delattre T, Baguette M, Burel F, Stevens VM, Quénol H, Vernon P (2013) Interactive effects of landscape and weather on dispersal. Oikos 122:1576–1585. doi:10.1111/j.1600-0706.2013.00123.x
Dennis RLH, Dapporto L, Dover JW, Shreeve TG (2013) Corridors and barriers in biodiversity conservation: a novel resource-based habitat perspective for butterflies. Biodivers Conserv 22:2709–2734. doi:10.1007/s10531-013-0540-2
Dover J, Settele J (2009) The influences of landscape structure on butterfly distribution and movement: a review. J Insect Conserv 13:3–27. doi:10.1007/s10841-008-9135-8
Dover J, Sparks T, Clarke S, Gobbett K, Glossop S (2000) Linear features and butterflies: the importance of green lanes. Agric Ecosyst Environ 80:227–242. doi:10.1016/S0167-8809(00)00149-3
Drag L, Hauck D, Pokluda P, Zimmermann K, Cizek L (2011) Demography and dispersal ability of a threatened saproxylic beetle: a mark-recapture study of the Rosalia Longicorn (Rosalia alpina). PLoS ONE 6:e21345. doi:10.1371/journal.pone.0021345
Dubois GF, Gouar PJ, Delettre YR, Brustel H, Vernon P (2010) Sex-biased and body condition dependent dispersal capacity in the endangered saproxylic beetle Osmoderma eremita (Coleoptera: Cetoniidae). J Insect Conserv 14:679–687. doi:10.1007/s10841-010-9296-0
Eriksson A, Low M, Berggren A (2013) Influence of linear versus network corridors on the movement and dispersal of the bush-cricket Metrioptera roeseli (Orthoptera: Tettigoniidae) in an experimental landscape. Eur J Entomol 110:81–86. doi:10.14411/eje.2013.010
Eycott AE, Stewart GB, Buyung-Ali LM, Bowler DE, Watts K, Pullin AS (2012) A meta-analysis on the impact of different matrix structures on species movement rates. Landsc Ecol 27:1263–1278. doi:10.1007/s10980-012-9781-9
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Annu Rev Ecol Evol Syst 34:487–515. doi:10.1146/annurev.ecolsys.34.011802.132419
Gillies CS, St Clair CC (2008) Riparian corridors enhance movement of a forest specialist bird in fragmented tropical forest. Proc Natl Acad Sci USA 105:19774–19779. doi:10.1073/pnas.0803530105
Goslee S, Urban D (2007) The ecodist package: dissimilarity-based functions for ecological analysis. J Stat Softw 22:1–19
Guerriero V, Vitale S, Ciarcia S, Mazzoli S (2011) Improved statistical multi-scale analysis of fractured reservoir analogues. Tectonophysics 504:14–24. doi:10.1016/j.tecto.2011.01.003
Haddad NM, Bowne DR, Cunningham A, Danielson BJ, Levey DJ, Sargent S, Spira T (2003) Corridor use by diverse taxa. Ecology 84:609–615. doi:10.1890/0012-9658(2003)084[0609:CUBDT]2.0.CO;2
Hardy OJ (2003) Estimation of pairwise relatedness between individuals and characterization of isolation-by-distance processes using dominant genetic markers. Mol Ecol 12:1577–1588
Hardy OJ, Vekemans X (1999) Isolation by distance in a continuous population: reconciliation between spatial autocorrelation analysis and population genetics models. Heredity (Edinb) 83:145–154. doi:10.1046/j.1365-2540.1999.00558.x
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620. doi:10.1046/j.1471-8278
Hedin J, Ranius T (2002) Using radio telemetry to study dispersal of the beetle Osmoderma eremita, an inhabitant of tree hollows. Comput Electron Agric 35:171–180. doi:10.1016/S0168-1699(02)00017-0
Horák J, Vávrová E, Chobot K (2010) Habitat preferences influencing populations, distribution and conservation of the endangered saproxylic beetle Cucujus cinnaberinus (Coleoptera: Cucujidae) at the landscape level. Eur J Entomol 107:81–88. doi:10.14411/eje.2010.011
Jonsson M (2003) Colonisation ability of the threatened tenebrionid beetle Oplocephala haemorrhoidalis and its common relative Bolitophagus reticulatus. Ecol Entomol 28:159–167. doi:10.1046/j.1365-2311.2003.00499.x
Jopp F, Reuter H (2005) Dispersal of carabid beetles—emergence of distribution patterns. Ecol Model 186:389–405. doi:10.1016/j.ecolmodel.2005.02.009
Jordán F, Magura T, Tóthmérész B, Vasas V, Ködöböcz V (2007) Carabids (Coleoptera: Carabidae) in a forest patchwork: a connectivity analysis of the Bereg Plain landscape graph. Landsc Ecol 22:1527–1539. doi:10.1007/s10980-007-9149-8
Kadej M, Zając K, Ruta R, Gutowski JM, Tarnawski D, Smolis A, Olbrycht T, Malkiewicz A, Myśków E, Larsson MC, Andersson F, Hedenström E (2015) Sex pheromones as a tool to overcome the Wallacean shortfall in conservation biology: a case of Elater ferrugineus Linnaeus, 1758 (Coleoptera: Elateridae). J Insect Conserv 19:25–32. doi:10.1007/s10841-014-9735-4
Koen EL, Garroway CJ, Wilson PJ, Bowman J (2010) The effect of map boundary on estimates of landscape resistance to animal movement. PLoS ONE 5:8. doi:10.1371/journal.pone.0011785
Kool JT, Moilanen A, Treml EA (2012) Population connectivity: recent advances and new perspectives. Landsc Ecol 28:165–185. doi:10.1007/s10980-012-9819-z
Kuefler D, Hudgens B, Haddad NM, Morris WF, Thurgate N (2010) The conflicting role of matrix habitats as conduits and barriers for dispersal. Ecology 91:944–950. doi:10.1890/09-0614.1
Larsson MC, Svensson GP (2009) Pheromone monitoring of rare and threatened insects: exploiting a pheromone-kairomone system to estimate prey and predator abundance. Conserv Biol 23:1516–1525. doi:10.1111/j.1523-1739.2009.01263.x
Le Gouar PJ, Dubois GF, Vignon V, Brustel H, Vernon P (2014) Demographic parameters of sexes in an elusive insect: implications for monitoring methods. Popul Ecol. doi:10.1007/s10144-014-0453-2
Lowe WH, Allendorf FW (2010) What can genetics tell us about population connectivity? Mol Ecol 19:3038–3051. doi:10.1111/j.1365-294X.2010.04688.x
Lozier JD, Strange JP, Koch JB (2013) Landscape heterogeneity predicts gene flow in a widespread polymorphic bumble bee, Bombus bifarius (Hymenoptera: Apidae). Conserv Genet 14:1099–1110. doi:10.1007/s10592-013-0498-3
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol 18:189–197. doi:10.1016/S0169-5347(03)00008-9
McRae BH (2006) Isolation by resistance. Evolution 60:1551–1561. doi:10.1554/05-321.1
McRae BH, Beier P (2007) Circuit theory predicts gene flow in plant and animal populations. Proc Natl Acad Sci USA 104:19885–19890. doi:10.1073/pnas.0706568104
Merckx T, Van Dyck H, Karlsson B, Leimar O (2003) The evolution of movements and behaviour at boundaries in different landscapes: a common arena experiment with butterflies. Proc Biol Sci 270:1815–1821. doi:10.1098/rspb.2003.2459
Michalcewicz J, Ciach M (2012) Rosalia longicorn Rosalia alpina (L.) (Coleoptera: Cerambycidae) uses roadside European ash trees Fraxinus excelsior L.—an unexpected habitat of an endangered species. Pol J Entomol 81:49–56. doi:10.2478/v10200-011-0063-7
Moilanen A, Hanski I (1998) Metapopulation dynamics: effects of habitat quality and landscape structure. Ecology 79:2503–2515. doi:10.1890/0012-9658(1998)079[2503:Mdeohq]2.0.Co;2
Musa N, Andersson K, Burman J, Andersson F, Hedenström E, Jansson N, Paltto H, Westerberg L, Winde I, Larsson MC, Bergman K-O, Milberg P (2013) Using sex pheromone and a multi-scale approach to predict the distribution of a rare saproxylic beetle. PLoS ONE 8:e66149. doi:10.1371/journal.pone.0066149
Nieto A, Alexander KNA (2010) European red list of saproxylic beetles. Publications Office of the European Union, Luxembourg
Öckinger E, Smith HG (2006) Semi-natural grasslands as population sources for pollinating insects in agricultural landscapes. J Appl Ecol 44:50–59. doi:10.1111/j.1365-2664.2006.01250.x
Öckinger E, Smith HG (2008) Do corridors promote dispersal in grassland butterflies and other insects? Landsc Ecol 23:27–40. doi:10.1007/s10980-007-9167-6
Oleksa A, Ulrich W, Gawroński R (2006) Occurrence of the marbled rose-chafer (Protaetia lugubris Herbst, Coleoptera, Cetoniidae) in rural avenues in northern Poland. J Insect Conserv 10:241–247. doi:10.1007/s10841-005-4830-1
Oleksa A, Ulrich W, Gawroński R (2007) Host tree preferences of hermit beetles (Osmoderma eremita Scop., Coleoptera: Scarabaeidae) in a network of rural avenues in Poland. Pol J Ecol 55:315–323
Oleksa A, Chybicki IJ, Gawroński R, Svensson GP, Burczyk J (2013a) Isolation by distance in saproxylic beetles may increase with niche specialization. J Insect Conserv. doi:10.1007/s10841-012-9499-7
Oleksa A, Tofilski A, Gawroński R, Tofilski A (2013b) Rural avenues as a refuge for feral honey bee population. J Insect Conserv 17:465–472. doi:10.1007/s10841-012-9528-6
Ouin A, Burel F (2002) Influence of herbaceous elements on butterfly diversity in hedgerow agricultural landscapes. Agric Ecosyst Environ 93:45–53. doi:10.1016/S0167-8809(02)00004-X
Oyler-McCance SJ, Fedy BC, Landguth EL (2013) Sample design effects in landscape genetics. Conserv Genet 14:275–285. doi:10.1007/s10592-012-0415-1
Pawłowski J, Kubisz D, Mazur M (2002) Coleoptera—chrząszcze. In: Głowaciński Z (ed) Czerwona lista zwierząt ginących i zagrożonych w Polsce. Instytut Ochrony Przyrody PAN, Kraków
Pradines C (2009) Road infrastructures: tree avenues in the landscape. 5th Council of europe conference on the european landscape convention. Document CEP-CDPATEP (2009) 15E. Council of Europe, Strasbourg
R Developement Core Team R (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Ranius T, Hedin J (2001) The dispersal rate of a beetle, Osmoderma eremita, living in tree hollows. Oecologia 126:363–370. doi:10.1007/s004420000529
Ries L, Debinski DM (2001) Butterfly responses to habitat edges in the highly fragmented prairies of Central Iowa. J Anim Ecol 70:840–852. doi:10.1046/j.0021-8790.2001.00546.x
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Rousset F (2000) Genetic differentiation between individuals. J Evol Biol 13:58–62. doi:10.1046/j.1420-9101.2000.00137.x
Row JR, Blouin-Demers G, Lougheed SC (2010) Habitat distribution influences dispersal and fine-scale genetic population structure of eastern foxsnakes (Mintonius gloydi) across a fragmented landscape. Mol Ecol 19:5157–5171. doi:10.1111/j.1365-294X.2010.04872.x
Schwartz MK, Copeland JP, Anderson NJ, Squires JR, Inman RM, McKelvey KS, Pilgrim KL, Waits LP, Cushman SA (2009) Wolverine gene flow across a narrow climatic niche. Ecology 90:3222–3232. doi:10.1890/08-1287.1
Skórka P, Lenda M, Moroń D, Kalarus K, Tryjanowski P (2013) Factors affecting road mortality and the suitability of road verges for butterflies. Biol Conserv 159:148–157. doi:10.1016/j.biocon.2012.12.028
Spear SF, Balkenhol N, Fortin M-J, McRae BH, Scribner K (2010) Use of resistance surfaces for landscape genetic studies: considerations for parameterization and analysis. Mol Ecol 19:3576–3591. doi:10.1111/j.1365-294X.2010.04657.x
Stamps JA, Krishnan VV, Reid ML (2005) Search costs and habitat selection by dispersers. Ecology 86:510–518. doi:10.1890/04-0516
Sturges H (1926) The choice of a class interval. J Am Stat Assoc 21:65–66
Svensson GP, Larsson MC (2008) Enantiomeric specificity in a pheromone-kairomone system of two threatened saproxylic beetles, Osmoderma eremita and Elater ferrugineus. J Chem Ecol 34:189–197. doi:10.1007/s10886-007-9423-x
Svensson GP, Larsson MC, Hedin J (2004) Attraction of the larval predator Elater ferrugineus to the sex pheromone of its prey, Osmoderma eremita, and its implication for conservation biology. J Chem Ecol 30:353–363. doi:10.1023/B:JOEC.0000017982.51642.8c
Svensson GP, Sahlin U, Brage B, Larsson MC (2011) Should I stay or should I go? Modelling dispersal strategies in saproxylic insects based on pheromone capture and radio telemetry: a case study on the threatened hermit beetle Osmoderma eremita. Biodivers Conserv 20:2883–2902. doi:10.1007/s10531-011-0150-9
QGIS Development Team (2012) QGIS geographic information system. Open Source Geospatial Foundation Project. http://qgis.osgeo.org.qgisorg
Tewksbury JJ, Levey DJ, Haddad NM, Sargent S, Orrock JL, Weldon A, Danielson BJ, Brinkerhoff J, Damschen EI, Townsend P (2002) Corridors affect plants, animals, and their interactions in fragmented landscapes. Proc Natl Acad Sci USA 99:12923–12926. doi:10.1073/pnas.202242699
Tolasch T, Von Fragstein M, Steidle JLM (2007) Sex pheromone of Elater ferrugineus L. (Coleoptera: Elateridae). J Chem Ecol 33:2156–2166. doi:10.1007/s10886-007-9365-3
Travis JMJ, Dytham C (1999) Habitat persistence, habitat availability and the evolution of dispersal. Proc R Soc B Biol Sci 266:723. doi:10.1098/rspb.1999.0696
van Etten J (2014) gdistance: distances and routes on grids. R package version 3.1.0. http://CRAN.R-project.org/package=gdistance
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13:921–935. doi:10.1046/j.1365-294X.2004.02076.x
Vekemans X, Beauwens T, Lemaire M, Roldán-Ruiz I (2002) Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol Ecol 11:139–151. doi:10.1046/j.0962-1083.2001.01415.x
von Schrötter FL (1812) Karte von Ost-Preussen nebst Preussisch Litthauen und West-Preussen nebst dem Netzdistrict. S. Schropp & Company, Berlin
Vos P, Hogers R, Bleeker M, Reijans M, Van De Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414. doi:10.1093/nar/23.21.4407
Walker MP, Dover JW, Sparks TH, Hinsley SA (2006) Hedges and green lanes: vegetation composition and structure. Biodivers Conserv 15:2595–2610. doi:10.1007/s10531-005-4879-x
Whitlock R, Hipperson H, Mannarelli M, Butlin RK, Burke T (2008) An objective, rapid and reproducible method for scoring AFLP peak-height data that minimizes genotyping error. Mol Ecol Resour 8:725–735. doi:10.1111/j.1471-8286.2007.02073.x
Williams WI, Robertson IC (2008) Using automated flight mills to manipulate fat reserves in Douglas-fir beetles (Coleoptera: Curculionidae). Environ Entomol 37:850–856. doi:10.1603/0046-225X(2008)37[850:UAFMTM]2.0.CO;2
Wright S (1943) Isolation by distance. Genetics 28:114–138
Zauli A, Chiari S, Hedenström E, Svensson GP, Carpaneto GM (2014) Using odour traps for population monitoring and dispersal analysis of the threatened saproxylic beetles Osmoderma eremita and Elater ferrugineus in central Italy. J Insect Conserv 18:801–813. doi:10.1007/s10841-014-9687-8
Zeller KA, McGarigal K, Whiteley AR (2012) Estimating landscape resistance to movement: a review. Landsc Ecol 27:777–797. doi:10.1007/s10980-012-9737-0
Acknowledgments
The study was supported by the National Science Centre (Grant No. UMO-2011/03/B/NZ9/03103) and by the Municipality of Rychliki. The samples were collected held with the permission of the General Directorate for Environmental Protection (permission no. DOPozgiz-4200/I-105/2674/10/JRO). The authors would like to thank Katarzyna Meyza and Ewa Sztupecka for their help with laboratory work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Oleksa, A., Chybicki, I.J., Larsson, M.C. et al. Rural avenues as dispersal corridors for the vulnerable saproxylic beetle Elater ferrugineus in a fragmented agricultural landscape. J Insect Conserv 19, 567–580 (2015). https://doi.org/10.1007/s10841-015-9778-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10841-015-9778-1