Abstract
Genetic variation of the globally threatened obligatorily myrmecophilous Large Blue butterfly Phengaris (Maculinea) arion (Lepidoptera) was studied, using six microsatellite markers, in a country where its decline is dramatic (Poland). Material was collected on 13 sites showing considerable ecological variation as far as biotope, larval food plant and host ants of the butterfly were concerned. Genetic variability, estimated in terms of number of alleles and heterozygosity, was the lowest in the most isolated populations. However on sites localized in areas where suitable biotopes were extensive and interconnected, P. arion still held relatively high genetic diversity. Pairwise F ST values indicated small and moderate differentiation among samples (F ST = 0.01–0.15), with the exceptions of two isolated localities (0.20). We did not find clear evidence of isolation by distance. The presence of four or five genetic clusters was indicated. Analysis of the membership of each individual to each cluster showed that the vast majority of individuals from three isolated populations were clustered in three separate genetic groups. The most distinct population was the one, which had been found to be specialized towards Myrmica lobicornis in previous studies. Individuals from the remaining populations could not be clustered in separate genetic groups, however some dominance of different clusters in geographical regions was observed. Some portion of the population’s genetic variability could be explained by geographical distribution, however the percentage of variation, explaining the differences between two main regions (S and NE Poland), was very low. We conclude that the main factor shaping the current genetic structure of P. arion in Poland is the recent isolation of populations related to habitat fragmentation but local ecological specializations may be also a potential factor. Therefore the necessity of activities aiming to halt the further reduction of genetic variability, as well as the monitoring of priority populations (e.g. those belonging to unique host races), should be emphasized in future action plans in Central Europe.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Butterflies (Lepidoptera: Hesperioidea, Papilionoidea) are charismatic taxa regarded as model organisms in studies of evolution, behaviour, ecology and biogeography (Settele et al. 2009). Many species also attract the attention of conservationists because they have experienced severe decline over the last few decades. However only some representatives of this group have achieved the status of icons in animal conservation and entered the canons of ecology. Palaearctic Large Blue Phengaris (Maculinea) arion is undoubtedly one of the best examples (Thomas and Settele 2004; Settele et al. 2005; Settele and Kühn 2009). The enormous interest in this species results from the dramatic decline, extinction, and then successful reintroduction of the butterfly in the UK (Thomas et al. 2009). The susceptibility of P. arion is related to a narrow and complex ecological niche of larvae requiring the coincidence of two different types of essential resources to complete their development. Caterpillars of P. arion are initially phytophagous, feeding on specific plants, but in their fourth (final) instar they turn into social parasites preying upon the Myrmica host brood in colonies of Myrmica ants. The first studies triggered by the disappearing of P. arion revealed that the survival rate of caterpillars in nests varies between ant species, and under UK conditions only an abundance of thermophilous M. sabuleti guarantees that the population will thrive in the long term (Thomas et al. 1989; Elmes et al. 1998). Restoration of habitats based on this acquired knowledge was followed by introductions of the butterfly. A recent increase in the number of sites and adults (Thomas et al. 2009) has made the UK trend exceptional compared to the rest of Europe (Van Swaay et al. 2010), where a decrease of P. arion in most countries has been observed. Agricultural abandonment or improvements as well as afforestation resulting in loss or at least isolation and fragmentation of habitat are reported as the main threats (Van Swaay and Warren 1999).
Until recently, studies on P. arion have concentrated on species ecology (e.g. Thomas 1995; Pauler-Fürste et al. 1996; Thomas et al. 1998; Thomas 2002; Mouquet et al. 2005; Spitzer et al. 2009; Casacci et al. 2010; Sielezniew et al. 2010a) and phylogeny, including molecular data (Als et al. 2004; Fric et al. 2007). However little is known about the genetic structure of the population at landscape, European or Palearctic level. Pecsenye et al. (2007) studied three populations of P. arion from eastern Central Europe separated by ca. 250–500 km, using allozymes, in comparison with their congenera. Genetic differentiation of P. arion, which could be ascribed as rather moderate, although significant, was higher than that observed for P. teleius and P. nausithous, but clearly lower than in the case of P. alcon populations. The low number of populations analysed made it impossible to characterize a wider pattern or the factors affecting genetic differentiation within the species. Recently Rutkowski et al. (2009) showed that microsatellite nuclear markers originally developed for congenera, i.e. P. alcon and P. nausithous (Zeisset et al. 2005), and used in the investigation of the molecular ecology of these species (Anton et al. 2007; Nash et al. 2008) may also be successfully applied in studies of P. arion. Preliminary data on the genetic structure of the butterfly in Poland also indicated moderate genetic differentiation among populations, but simultaneously suggested that some pairs of populations could be highly differentiated, despite spatial (geographical) closeness (Rutkowski et al. 2009).
In the present contribution we have investigated patterns of genetic differentiation of P. arion across the species range in a country where its decline is dramatic (Poland). The species has disappeared from western localities within the last few decades, and some extant populations thrive in fragmented habitat. However, it is still widespread in the south-eastern half of Poland and recorded at a latitudinal gradient from south to north over a few hundred kilometres (Sielezniew et al. 2005). In this area P. arion shows considerable ecological variation as far as biotope, larval food plant and host ants are concerned. In most of Poland the butterfly inhabits sandy biotopes (forest-steppes or clearings and road verges in dry pine forests) and uses Thymus serpyllum. Only in foothills and mountainous areas, P. arion is related to xerothermal grasslands (with T. pulegioides), which are more typical biotopes of the species on a European scale. Recent studies from Poland have also proved that at least some populations in this part of Europe do not depend on M. sabuleti, as larvae of P. arion were observed in the nests of six other species, showing a complicated pattern of host ant use (Sielezniew and Stankiewicz 2008; Sielezniew et al. 2010a, 2010b and 2010c).
Therefore we were interested in whether ecological variation of P. arion was followed by genetic differentiation, as observed for e.g. Euphydryas aurinia, where host plant use affects genetic structure (Descimon et al. 2001; Nève 2009). The alternative hypothesis was that isolation and geographical distance between sampling localities were more important factors. We also aimed to assess whether some populations could be identified as separate conservation units on the basis of ecological and molecular data (e.g. Vila et al. 2006). Since P. arion does not show variation in mitochondrial DNA in Poland (Sielezniew et al. unpublished) and application of allozymes was not possible for conservation and practical reasons, we used microsatellites as a tool.
Materials and methods
Localities and sampling
Material analysed in the present study was collected at 13 sites of P. arion in southern and eastern Poland i.e. throughout its present distribution range in the country (Sielezniew et al. 2005). Distance among localities ranged from 8 to 478 km. Details are given in Table 1 and the distribution of studied populations is also shown on a map (Fig. 1). Sampling was carried out between 2005 and 2008. The butterfly is univoltine and its flight period in Poland lasts from early June to early August with considerable variation concerning the peak, related to biotope and seasons. We obtained the total number of samples from 233 imagoes in two ways: (1) a specimen with heavily worn wings (preferably a male) i.e. hypothetically at the end of its life, was caught and placed into 96% alcohol, then a fragment of thorax was used as a source of DNA or (2) a single middle leg was removed and the butterfly was then immediately released. From every population we obtained 13–26 samples with the exception of one very small and vulnerable population from Truskaw (TRU) where we collected legs from only seven individuals. In most cases sampling was conducted in a very restricted area with a diameter of a few hundred metres. This was impossible only at Łużany (LUZ) and Orchówek (ORC), where butterflies were encountered in low densities but within the space of kilometres along forest roads, electricity lines and railways. Therefore at both these localities we sampled throughout the wider area of the continuous biotope of the butterfly.
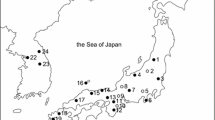
Location of the sampling sites of P. arion in Poland (for full names see Table 1), and proportion of membership of individuals from each predefined population in each of the four clusters (I, II, III, IV) indicated by the Structure analysis. The sizes of circles are proportional to the number of individuals analyzed. The TRU population was excluded because of too small sample size (7). The dotted line represents the western border of the present distribution range of P. arion in the country
To elucidate a pattern and the factors affecting the genetic structure of the species, populations were divided into groups reflecting geographical distribution or ecological variation (Table 1, see also Fig. 1). Four different grouping patterns were applied. (1) Geographical distribution on a regional scale i.e. seven regions including 1–5 populations were distinguished. (2) Geographical distribution on a macro-scale i.e. samples were divided into two groups only: NE Poland (eight populations) and S Poland (five populations). (3) Host plants (related to the type of biotope) i.e. ten and three populations using T. serpyllum (sandy and rather flat habitats) and T. pulegioides (xerothermal meadows on south exposed slopes) respectively. (4) Host ants i.e. three groups of populations where these relationships were studied. The first group included only one population, for which the specialisation to M. lobicornis was proven. The second group consisted of two populations (HUT and KLU) where M. sabuleti was found as a host ant. Finally, five populations, which all use M. schencki, and some of them also M. hellenica and/or M. rugulosa made up the third group. M. sabuleti was rare or absent at all those sites (Sielezniew and Stankiewicz 2008; Sielezniew et al. 2010a, 2010b and unpublished). We excluded from the analysis the SOS population, for which multiple host ant use is reported i.e. larvae/pupae of P. arion were found in nests of M. sabuleti, M. lobicornis, M. schencki and M. lonae (Sielezniew et al. 2010c).
Laboratory procedures
DNA was extracted from approximate 0.5 cm long fragments of thorax or from the whole leg, as described in Rutkowski et al. (2009). Using the PCR reaction we amplified six microsatellite loci, designated by Zeisset et al. (2005) as Macu8, Macu9, Macu11, Macu15, Macu16 and Macu17 using primers described by the authors. Amplification was performed using reagents and conditions described by Rutkowski et al. (2009).
The length of the amplified fragments, hence identification of microsatellite alleles, was estimated using a CEQ8000 Beckman Coulter automated sequencer (Comesa, Warsaw, Poland). Data were analysed using Beckman Coulter Fragment Analysis Software.
Statistical analysis
Analyses of the molecular data obtained were performed on a few different levels. First, we described microsatellite polymorphism for the whole studied Polish population of the species. In this analysis we estimated allelic diversity (A), observed heterozygosity (H O) and unbiased expected heterozygosity (H E) (Nei and Roychoudhury 1974) for each locus, using GenAlEx version 6.0 (Paekall and Smouse 2001) and FSTAT version 2.9.3 (Goudet 2001). We applied two tests for departures from Hardy–Weinberg Equilibrium (HWE): the first one took into account both heterozygote excess or deficit (probability test) and the second one took into account only heterozygote deficit. We tested departures from HWE for each of the six loci, as well as for all loci using Genepop on the Web version 4.0.10 (Raymond and Rousset 1995; Rousset 2008). Additionally, a fixation index (F IS) for each locus was calculated and its significance was tested under 1,680 randomisation using the Benjamini-Hochberg procedure (B–H) for controlling the false positive error rate in multiple comparison (Benjamini and Hochberg 1995), as implemented by Thissen et al. (2002). We also calculated the probability of genotypic linkage disequilibrium between all the pairs of loci within the species and applied the B–H method to control for multiple comparison again.
Secondly, we analysed microsatellite polymorphism within particular populations. Again, we estimated basic indicators of the population’s genetic variability (A, H O , H E), as well as allelic richness (R) and a mean number of “private” alleles (PA) for each population, and applied tests for departures from HWE, as described above. Indicators of genetic variability (mean A, R, and H O) among pairs of populations were compared with the Wilcoxon test using Statistica 6.0 software. We also calculated F IS values for each loci within each population, as well as overall F IS, and tested their significance under 1,560 permutations and B–H correction. Because the previous study (Rutkowski et al. 2009) shows that some microsatellite loci used in the present study may bear “null” alleles, with Macu16 as the most prone to this problem, we performed a test for the presence of null alleles within populations, using Micro-Checker (van Oosterhout et al. 2004). This analysis clearly showed that majority of populations had “null” alleles in locus Macu16, with a rather high frequency, thus we decided to exclude this locus from further analysis—all further calculations of genetic structure and genetic variation were performed using five microsatellite markers.
We estimated genetic differentiation among populations using a variety of methods. Overall and pairwise F ST (Weir and Cockerham 1984) estimates of genetic differentiation were obtained from FSTAT. Significance of pairwise F ST was tested under 1,560 permutation and with the B–H procedure. Also, for overall F ST we estimated 95% confidence intervals as implemented in FSTAT. To depict the results of pairwise F ST analysis a dendrogram based on a matrix of F ST values was prepared. We used MEGA 4 software (Tamura et al. 2007) to construct the Neighbour-joining (NJ) tree. The significance of correlation between genetic distance, defined as F ST/(1−F ST), and geographical distance was tested using the Mantel test implemented in FSTAT. Bayesian-clustering method (Structure version 2.3.2; Pritchard et al. 2000) was used to examine how well predefined ‘populations’ corresponded to genetic groups (K). We ran Structure three times for each user-defined K (1–12) with an initial burn in of 50 000 and 100 000 iterations of the total data set. We used the admixture model of ancestry, and the correlated model of alleles frequencies. The proportion of membership of each predefined ‘population’ within each genetic group was estimated for K with the highest likelihood. Sampling location was not used as prior information. Additionally, we performed the Structure analysis for five populations from NE Poland, which formed, unlike the others, a distinct regional group. In this analysis we defined K from 1 to 5 and applied the same conditions described above.
Next, we analysed microsatellite polymorphism and genetic structure taking into consideration predefined groups of populations. For each group we calculated basic indicators of genetic variability with GenAlEx and FSTAT. Using the AMOVA procedure in the Arlequin software, version 2.0 (Schneider et al. 2000) we estimated the proportion of variance among and within groups of populations. Stepwise mutation model (SMM) was assumed for the evolution of microsatellite markers. The significance of the observed variance components was evaluated by means of a non-parametric permutation method (Excoffier et al. 1992) as executed in Arlequin.
Results
Genetic diversity
The six microsatellite loci were polymorphic in all populations. A total number of 115 alleles were detected, ranging from eight (Macu16) to 29 (Macu9) per locus. In the analysis of the whole Polish population none of the loci were in HWE, as they showed significant F IS values in 5 out of 6 loci. In the analysis on a population level in six locations (PIA, LUZ, ORC, BAB, KLU, SRO) we found significant heterozygote deficiency in Macu16 and the HWE test showed that five of them (PIA, LUZ, ORC, BAB and SRO) were not in HWE. When we excluded from our analysis Macu16, for which the presence of null alleles was detected (see Material and Methods), all populations met HWE. Meanwhile, we found heterozygote deficiency, indicated by significant F IS, when grouping populations in regional and ecological groups (Table 2), which was most probably interlinked with the ‘Wahlund effect’ (clustering populations with restricted gene flow among them).
The test for linkage disequilibrium between all pairs of loci indicated significant results (after B–H correction for multiple comparisons) for two pairs of loci Macu15/Macu17 and Macu16/Macu17, however there was no significant linkage disequilibrium when each population was analysed separately.
The level of microsatellite polymorphism, in terms of number of alleles (A) and allelic richness (R), was the lowest in the case of GUG, TRU and HOR. In the GUG and HOR populations no “private” alleles were also found. The highest level of microsatellite polymorphism was detected for HUT, SUK, LUZ and SRO (Table 2). There were significant differences in pairwise analyses in 33 of 78 cases for A and in 27 of 78 cases for R e.g. in HUT the mean number of alleles and allelic richness was higher than in all other populations except of PIA, LUZ, SUK and SRO; and in GUG A and R were significantly lower than in the other seven and six populations respectively.
The observed heterozygosity ranged from 0.471 (GUG) to 0.830 (HUT) (Table 2). However, pairwise analyses showed significant differences (Wilcoxon test, P < 0.05) only in nine out of 78 cases i.e. GUG < HUT, GUG < KLU, GUG < SRO, HUT > SOW, HUT > LUZ, HUT > HOR, HUT > TRU, HUT > ORC and HOR < SUK.
When individuals were divided into two groups reflecting geographical distribution, mean values of A, R, H O were higher for S Poland than for NE Poland but differences were not significant (P = 0.5, P = 0.35, P = 0.08 respectively). When comparing groups of populations using different larval food plants and larval host ants, the only significant difference concerned A, which was higher for T. serpyllum than for T. pulegioides, and higher for M. schencki than for M. sabuleti (P < 0.05), respectively (Table 2). Other comparisons concerning host ants were not performed as there were only single populations, which used M. schencki and M. sabuleti simultaneously, or specialized to M. lobicornis.
Genetic population structure
The overall F ST was 0.073 (CI 95% = 0.065–0.083). All pairwise F ST values were significant after B–H correction, with the exception of PIA versus LUZ (F ST = 0.011). The majority of pairwise F ST values (71 of 78) showed moderate genetic differentiation (F ST ranged from 0.054 to 0.147). The highest value was found for GUG versus KLU (F ST = 0.201). The NJ tree of pairwise F ST values suggested some (rather weakly pronounced) pattern of differentiation on the north–south transect (Fig. 2). However the effect of isolation by distance was not found for the studied populations of P. arion (Mantel test: r2 = 0.08, P = 0.41) (Fig. 3).
Neighbour-joining tree based on pairwise F ST values among 13 populations of P. arion (for full names of the locations see Table 1)
The cluster analysis of the total sample in Structure showed the highest likelihoods for K = 4 and K = 5 (Fig. 4a). As far as four clusters are concerned (Fig. 4b), the pattern was the clearest: individuals from predefined populations i.e. GUG, HOR and KLU were clustered in three separate clades (II, III, I respectively). Individuals from the remaining populations were not clustered in separate clades.
Structure analysis of 226 individuals of P. arion from 12 sites (TRU excluded because of small sample size). a Scatter plot showing the estimated likelihood of each number of inferred genetic clusters. The third value for K = 8 was very low (−23,847) and is out of the scale. The optimal number of clusters, K = 4 and K = 5 were determined by highest log-likehood value and the lowest amount of variance for three independent iterations. b Bayesian assignment of individuals to four genetic groups (for K = 5 the pattern was less clear and therefore is not shown). Each bar represents the estimated posterior probability of each individual butterfly belonging to each of the four inferred clusters. Solid black lines define the boundaries between the populations, used in the analysis (see Table 1 and Fig. 1)
The proportion of individuals from separate populations clustered in genetic groups differed depending on the group. Three distinct clusters (I, II and III) contained respectively 90% of the individuals from the GUG population, 78.2% from the HOR population and 77.7% from the KLU population, whereas individuals from the majority of other predefined populations were distributed more or less evenly among all four clusters, especially in case of LUZ (16.0–30.1%), ORC (18.2–36.2%), SOW (13.7–34.7%), SUK (19.1–37.9%) and BAB (14.0–38.4%) (Fig. 1). However, it could be stated that two clusters (II and III) dominated in NE Poland, whereas cluster IV dominated in the highlands and foothills of S Poland; and the majority of individuals from two mountainous populations were clustered to the clade I. In the case of five clusters (not shown) only GUG and KLU consisted of individuals from single clusters.
When we restricted our analyses to samples collected in one region i.e. five localities from Podlasie (SOS, GUG, PIA, SOW, LUZ), the optimal number of clusters was K = 3 (Fig. 5a). All individuals from GUG were clustered separately, whereas individuals from the other four populations were distributed among two other clusters. However two individuals from PIA and one individual from SOS had the highest likelihood of membership in the ‘GUG cluster’ (Fig. 5b). Pairwise F ST comparisons among five populations from the region suggested the puzzling distinctness of GUG, e.g. values were lower (0.011–0.067) for four populations (without GUG) than when GUG were set against them (0.101–0.124).
Structure analysis of 93 individuals of P. arion from five sites concentrated in one region of NE Poland (Podlasie). a Scatter plot showing the estimated likelihood of each number of inferred genetic clusters. The optimal number of clusters, K = 3 were determined by highest log-likehood value and the lowest amount of variance for three independent iterations. b Bayesian assignment of individuals to three genetic groups. Each bar represents the estimated posterior probability of each individual butterfly belonging to each of the three inferred clusters. Solid black lines define the boundaries between the populations, used in the analysis (see Table 1 and Fig. 1)
Analysis of molecular variance for groups of populations formed due to geographical distribution (regional scale and macro-scale), or ecological differentiation (larval food plants and host ants), indicated that most of the variation might be explained by variation within a population (Vc = 91.24–92.56%) and among population within groups (Vb = 5.89–7.36%), and all of those values were highly significant (P < 0.0001). However we found that the division of samples into two main groups (NE Poland and S Poland) also gave significant results as far as Va (1.93%) is concerned, (F CT = 0.01935, P < 0.001) contrasting with more detailed grouping of samples from localities from the same regions (Va = 0.08, P = 0.42). Division of populations into two groups reflecting difference in host plant and host ant use explained 1.15% (F CT = 0.01147, P = 0.06) and 2.86% (F CT = 0.02857, P = 0.045) of variance respectively. However when GUG–the most isolated population—was excluded the latter grouping did not yield a significant value of Va component either.
Overall F ST values calculated for groups of populations inhabiting different biotopes and using different larval food plants (xerothermal meadows with T. pulegioides versus sandy habitats with T. serpyllum) were similar (0.068 [95% CI 0.045–0.087] and 0.079 [95% CI 0.053–0.107] respectively), and the difference was not statistically significant (one-sided test, P > 0.05).
Discussion
Our study elucidated genetic variability within populations and differentiation among Polish populations of Phengaris arion. All analysed microsatellite loci were highly polymorphic, which confirms the usefulness of these cross-amplified microsatellites in the population genetic study of this butterfly species. The genetic variability of P. arion seems to be relatively high compared to two congenera i.e. P. nausithous where allelic richness per population varies from 3.5 to 5.5 (Anton et al. 2007) and especially to P. ‘rebeli’ where allelic richness is: 1.8–2.2 (Rutkowski et al. 2009). This is also consistent with data of Pecsenye et al. (2007) on allozymes.
We found significant differences in the level of microsatellite polymorphism among studied populations of P. arion in Poland. This could be interlinked with differences in a level of within-population genetic variability. Effective population size is considered one of the main factors influencing genetic variability. Indeed, data obtained from populations of some other lycaenid butterflies with limited mobility indicate that genetic diversity is positively related to population size (Brookes et al. 1997; Harper et al. 2003). Unfortunately neither present nor historical detailed data on population census were available except for some general impressions and estimations.
Isolation is another factor influencing genetic variability, especially in species with limited dispersal abilities. It is predicted that in isolated populations allelic variation is declining faster than heterozygosity (Maruyama and Fuerst 1985). Indeed, in Polish populations of P. arion differences in genetic variability, estimated based on the number of microsatellite alleles and heterozygosity, were generally more pronounced in the case of number of alleles than in the level of heterozygosity. Undoubtedly, at least some of the studied populations were spatially isolated, and genetic drift had already started to eliminate some microsatellite alleles, and possibly some portion of population genetic variability. Reduced genetic variability increases extinction risk in butterflies because of lowered adaptability (Saccheri et al. 1998; Nieminen et al. 2001; Schmitt and Hewitt 2004), which e.g. for Polyommatus coridon results in a decrease of adult lifetime expectancy, and it is also negatively correlated with the number of dispersing individuals (Vandewoestijne et al. 2008). We suggest that our data indicate the necessity of active protection of the species in Poland, at least in the case of some populations, aiming to halt further reduction of genetic variability.
In the present study the highest level of microsatellite polymorphism was observed for three populations from S Poland (HUT, SUK, SRO), and one from the NE (LUZ). All of them were localised in areas where biotopes suitable for P. arion were extensive (at least in the recent past) and the butterfly was widespread, although not usually recorded in high numbers. HUT, SUK and LUZ populations inhabited sandy and mostly flat areas in dry pine forests, often of a ‘corridor’ character i.e. roadsides or clearings (including very long ones under electricity lines), with borders therefore not easy to determine. In HUT and SUK all samples were collected in a restricted area, but in LUZ because of the low density of adults sampling was performed over an extended area with a diameter of about 6 km. The relatively high value of F IS (among all studied populations only GUG has a higher F IS), however non-significant after B–H correction, could therefore result from the Wahlund effect. Almost nothing is known about the dispersal abilities of P. arion in such types of habitats, but our findings suggest the existence of small-scale genetic structure even in continuous habitat. The species is generally considered as sedentary, although studies carried out in Germany on xerothermal meadows also recorded the movements of single females over a distance of up to nearly 2 km, and also across forest (Pauler-Fürste et al. 1996). There are examples of small-scale genetic structure in Lepidoptera populations, induced by heterogeneous landscape, both in ‘low gene flow’ (e.g. Wynne et al. 2003) and ‘high gene flow’ species (e.g. Keyghobadi et al. 2005). Although the LUZ population inhabited a relatively homogenous biotope of dry forest, the habitat of the butterfly was in fact fragmented due to the patchy distribution of the host plant and, especially, the host ant species. However this hypothesis requires further, more detailed study of that population, as well as other P. arion populations inhabiting large areas. Interestingly, the other population sampled over a large area, i.e. ORC, had a smaller, but still positive F IS (however non-significant after correction).
The fourth genetically rich population (SRO) inhabited the Pieniny Mts. At the time of study the butterfly was not abundant and local there, but it used to be quite a widespread and common species just a few decades ago. In spite of recent decline the population seemed to retain its genetic diversity. Lower, though insignificantly, values of A and R were observed in nearby KLU where a numerous but isolated population existed. It also possessed lower PA than SRO, which may suggest that some rare alleles have just begun to be eliminated from this population, probably due to isolation and genetic drift. Despite geographical proximity we found significant genetic differentiation between SRO and KLU, confirming the isolation of the latter. Moreover, the Structure analysis suggests that the KLU population is the most homogenous of all those studied in S Poland. The distance between both KLU and SRO localities was about 8 km, however the localities were situated in different mountain ranges and also separated by dense coniferous forest and built-up areas. There are also slight phenological differences limiting gene flow i.e. the flight period in KLU is somewhat delayed compared to SRO, due to difference in altitude. Artificial exchange of imagoes should be considered in future action plans, since the creation of corridors between populations is hardly feasible. Ecological data suggest that both populations depend on the same ant species i.e. M. sabuleti (Sielezniew et al. 2010a; Sielezniew unpublished).
The lowest values of polymorphism observed for GUG, TRU and HOR may be explained well by the clear isolation of those populations. In the case of HOR there were no potential habitats in the radius of about 15 km and the nearest known P. arion site was about 25 km away, separated by agricultural land and forests. The TRU population was a very small one situated at the western edge of the species range in Poland and it was rediscovered only in the third year of thorough inventory work. Additionally, a negative value of F IS in this population might be a ‘genetic track’ of a recent bottleneck or a founder effect, although the small sample size makes any conclusions speculative.
The GUG population showed to be clearly distinct as indicated both by pairwise F ST comparisons and the Structure analysis. Surprisingly it was localised in the region where P. arion was not a rare species and neighbouring populations were about 10 km away at most. However the site was surrounded by wet biotopes which apparently acted as an effective barrier compared to mostly dry areas of forests and extensive agriculture. Interestingly, according to the Structure analysis restricted to NE Poland, a few specimens, which were clustered with the highest likelihood to the ‘GUG cluster’, were found among individuals collected from neighbouring populations i.e. PIA and SOS. It suggested that they could be the offspring of migrants from the original population and therefore there could be some appropriate habitats for P. arion between GUG and two other localities, supporting a stepping-stone gene flow similar to that observed for lycaenid Euphilotes enoptes (Peterson 1996). However the homogenous character of the GUG population suggests an asymmetrical gene flow, which might be explained by the landscape structure i.e. the low probability of finding a small isolated patch of habitat (1.3 ha) at GUG by a dispersing butterfly from an another locality.
The GUG population was also quite unique as it was the only known population of P. arion where local specialisation towards any host ant was proven in the field—i.e. the rare M. lobicornis is used but not the common M. sabuleti (Sielezniew et al. 2010a). Combined molecular and ecological data did not answer the question of conservation recommendations for this population. The specialisation in host ant relationships could result from isolation. At SOS—one of the two neighbouring localities—both M. lobicornis and M. sabuleti were recorded as hosts of the butterfly (Sielezniew et al. 2010c). Unfortunately at the PIA locality M. lobicornis is absent and M. sabuleti is a very rare ant (Sielezniew and Stankiewicz 2008), hence drawing further conclusions from ecological data is impossible. However we would rather suggest conservation (and monitoring) of this unique host race existing at GUG rather than e.g. introducing individuals from neighbouring localities to increase genetic diversity. Significant differentiation was also recorded between two populations (SOW and HOR) which were very similar as far as biotope and host ant use (exclusively M. schencki) are concerned (Sielezniew et al. 2010b), which implied that ecological and genetic variability did not overlap.
Genetic differentiation among populations of sedentary species of butterflies whose habitat is specific, and highly fragmented, is higher than for mobile generalists (Louy et al. 2007 and references therein). We found small yet significant genetic structure in the Polish population of P. arion. The overall F ST value of 0.073 is slightly lower than the one reported previously by Rutkowski et al. (2009) for P. arion populations in Poland. However this is a preliminary study based on a smaller number of individuals and populations sampled. The only other studies on population differentiation among populations of P. arion implemented allozymes and were performed in Hungary and Slovenia (F ST = 0.097), but only three populations (44 individuals) were analysed (Pecsenye et al. 2007). Some authors suggest that microsatellite markers give similar or smaller F ST values than allozymes (Meglécz et al. 1998; Sigaard et al. 2008 and references therein), but direct comparisons should be made with care.
The studies on genetic differentiation of butterflies are focused on rare and threatened sedentary species (Nève 2009). This bias is especially emphasised for analyses using microsatellite markers, as data concerning common and mobile species are barely available. Therefore it is not surprising that our calculation of overall differentiation fell within the range of results of other authors studying threatened butterflies in Europe (see Nève 2009 for a review; Nash et al. 2008; Sigaard et al. 2008; Finger et al. 2009 and Rutkowski et al. 2009). However it is worth noting that P. arion could rather be placed into the group of species with relatively lower differentiation.
On the other hand, for some pairs of populations we found quite high values of F ST, with the striking example of two very isolated populations: GUG and KLU (F ST = 0.201). Hence, spatial isolation of populations by unsuitable habitat seems to be an important factor in shaping the genetic structure of P. arion in Poland. We did not find evidence for isolation by distance, which was observed, for example, by Anton et al. (2007) in P. nausithous—a close relative of P. arion. However this study was performed on many populations (34) and on a much more restricted geographical scale (30 × 45 km). It is known that the effect of genetic differentiation may be clear at short distances but insignificant at longer distances because of genetic drift and adaptations to local conditions (Hutchinson and Templeton 1999). This kind of pattern was revealed for P. bellargus (Harper et al. 2003) and in other low-dispersing invertebrates, for example snails (Schweiger et al. 2004). In our study the overall lower number of populations sampled did not enable us to form ultimate conclusions about whether this phenomena was also applicable for P. arion. However, the relatively high values of genetic differentiation between populations situated in close geographical proximity suggest that landscape connectivity might be much more important factor than isolation by distance.
The AMOVA analysis did not indicate any clear pattern of hierarchical genetic structure in the Polish population of P. arion. The amount of genetic variance among groups of populations selected for geographical distribution or ecological variation was lower than within those groups. The only significant values of this component were found for division into two main regions, i.e. S and NE Poland (maybe also suggesting the relevance of isolation by distance), and for grouping according to host ants. However the latter results were biased by the distinctness of GUG population, which formed one of three groups. Contrastingly studies of Harper et al. (2003) on P. bellargus in UK, also with the application of microsatellite markers, revealed variation among geographical regions double that of the variation observed within regions. This may be explained by the status of the butterfly, which is not only sedentary but whose populations have been isolated by habitat fragmentation for a long time.
Although some of the genetic variability in the Polish population of P. arion could be explained by genetic differences between two geographical regions, which is also supported by the analysis in Structure and visible in the NJ tree, we rather conclude that the current genetic structure of P. arion in Poland is probably better explained by past distribution, when the butterfly used to be much more widespread and common, than by present distribution, which is strongly influenced by habitat fragmentation. Orsini et al. (2008) found such a kind of relationship at the metapopulation level for nymphalid Melitaea cinxia. The decline of P. arion in Poland is a quickly ongoing process (Sielezniew et al. 2005) and therefore many local populations which could contribute to a stepping-stone gene flow between those presently studied could have become extinct in the recent past. The past long distance gene flow is still visible in the Structure analysis and the differentiation between populations observed resulted rather from the recent genetic drift. To test this hypothesis it would be interesting to compare the genetic diversity of extant populations with museum specimens, which are quite numerous in Polish collections. The results of Harper et al. (2006) who successfully investigated century old material from another lycaenid species indicate the relevance and promise of such intentions.
References
Als TD, Vila R, Kandul NP, Nash DR, Yen Hsu, Mignault AA, Boomsma JJ, Pierce NE (2004) The evolution of alternative parasitic life histories in large blue butterflies. Nature 432:386–390
Anton C, Zeisset I, Musche M, Durka W, Boomsma JJ, Settele J (2007) Population structure of a large blue butterfly and its specialist parasitoid in a fragmented landscape. Mol Ecol 16:3828–3838
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc (B) 57:289–300
Brookes MJ, Graneau YA, King P, Rose OC, Thomas CS, Mallet JLB (1997) Genetic analysis of founder bottlenecks in the rare British butterfly Plebejus argus. Conserv Biol 11:648–661
Casacci LP, Witek M, Barbero F, Patricelli D, Solazzo G, Balletto E, Bonelli S (2010) Habitat preferences of Maculinea arion and its Myrmica host ants: implications for habitat management in Italian Alps. J Insect Conserv Published online first: 17 August 2010. DOI: 10.1007/s10841-010-9327-x
Descimon H, Zimmermann M, Cosson E, Barascud B, Nève G (2001) Diversité génétique, variation géographique et flux géniques chez quelques. Lépidoptères Rhopalocères français. Genet Sel Evol 33(Suppl 1):223–249
Elmes GW, Thomas JA, Wardlaw JC, Hochberg ME, Clarke RT, Simcox DJ (1998) The ecology of Myrmica ants in relation to the conservation of Maculinea butterflies. J Insect Conserv 2:67–78
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Finger A, Schmitt T, Meyer M, Assmann T, Zachos FE, Habel JC (2009) The genetic status of the Violet Copper Lycaena helle, a relict of the cold past in times of global warming. Ecography 32:382–390
Fric Z, Wahlberg N, Pech P, Zrzavý J (2007) Phylogeny and classification of the Phengaris-Maculinea clade (Lepidoptera: Lycaenidae): total evidence and phylogenetic species concepts. Syst Entomol 32:558–567
Goudet J (2001) FSTAT V2.9.3, a program to estimate and test gene diversities and fixation indices. http://www.unil.ch/izea/softwares/fstat.htlm
Harper GL, Goulson D, Maclean N (2003) Microsatellite markers to assess the influence of population size, isolation and demographic change on the genetic structure of the UK butterfly Polyommatus bellargus. Mol Ecol 12:3349–3357
Harper GL, Maclean N, Goulson D (2006) Analysis of museum specimens reveals extreme genetic drift in the Adonis Blue butterfly (Polyommatus bellargus). Biol J Linn Soc 88:447–452
Hutchinson DW, Templeton AR (1999) Correlation of pairwise genetic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 53:1898–1914
Keyghobadi N, Roland J, Strobeck C (2005) Genetic differentiation and gene flow among populations of the alpine butterfly, Parnassius smintheus, vary with landscape connectivity. Mol Ecol 14:1897–1909
Louy D, Habel JC, Schmitt T, Meyer M, Assmann T, Müller P (2007) Strongly diverging population genetic patterns of three skipper species: isolation, restricted gene flow and panmixis. Conserv Genet 8:671–681
Maruyama T, Fuerst PA (1985) Population bottlenecks and nonequilibrium models in population genetics II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 111:675–690
Meglécz E, Pecsenye K, Varga Z, Solignac M (1998) Comparison of differentiation pattern at allozyme and microsatellite loci in Parnassius mnemosyne (Lepidoptera) populations. Hereditas 128:95–103
Mouquet N, Thomas JA, Elmes GW, Clarke RT, Hochberg ME (2005) Population dynamics and conservation of a specialized predator: a case study of Maculinea arion. Ecol Monogr 75:525–542
Nash DR, Als TD, Maile R, Jones GR, Boomsma JJ (2008) A mosaic of chemical coevolution in a large blue butterfly. Science 319:88–90
Nei M, Roychoudhury AK (1974) Sampling variances of heterozygosity and genetic distance. Genetics 76:379–390
Nève G (2009) Population genetics of butterflies. In: Settele J, Shreeve T, Konvička M, Van Dyck H (eds) Ecology of butterflies in Europe. Cambridge University Press, Cambridge, pp 107–129
Nieminen M, Singer MC, Fortelius W, Schops K, Hanski I (2001) Experimental evidence that inbreeding increase extinction risk in butterfly populations. Am Nat 157:237–244
Orsini L, Jorander J, Alasentie A, Hanski I (2008) Genetic spatial structure in a butterfly metapopulation correlates better with past than present demographic structure. Mol Ecol 17:2629–2642
Paekall R, Smouse PE (2001) GenAlEx V5: Genetic analysis in excel. Population genetic software for teaching and research. http://www.anu.ed.au/BoZo/GenAlEx/
Pauler-Fürste R, Kaule G, Settele J (1996) Aspects of the population vulnerability of the large blue butterfly Glaucopsyche (Maculinea) arion in south-west Germany. In: Settele J, Margules C, Poschlod P, Henle K (eds) Species survival in fragmented landscapes. Kluwer, Dordrecht, pp 275–281
Pecsenye K, Bereczki J, Tihanyi B, Toth A, Peregovits L, Varga Z (2007) Genetic differentiation among the Maculinea species (Lepidoptera: Lycaenidae) in eastern Central Europe. Biol J Linn Soc 91:11–21
Peterson MA (1996) Long distance gene flow in the sedentary butterfly Euphilotes enoptes (Lepidoptera: Lycaenidae). Evolution 50:1990–1999
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Res 8:103–106
Rutkowski R, Sielezniew M, Szostak A (2009) Contrasting levels of polymorphism in cross-amplified microsatellites in two endangered xerothermophilous, obligatorily myrmecophilous, butterflies of the genus Phengaris (Maculinea) (Lepidoptera: Lycaenidae). Eur J Entomol 106:457–469
Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I (1998) Inbreeding and extinction in a butterfly metapopulation. Nature 392:491–494
Schmitt T, Hewitt GM (2004) The genetic pattern of population threat and loss: a case study of butterflies. Mol Ecol 13:21–31
Schneider SD, Roessli D, Excoffier L (2000) Arlequin ver. 2000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland
Schweiger O, Frenzel M, Durka W (2004) Spatial genetic structure in metapopulation of the land snail Cepaea nemoralis (Gastropoda: Helicidae). Mol Ecol 13:3645–3655
Settele J, Kühn E (2009) Insect conservation. Science 325:41–42
Settele J, Kühn E, Thomas JA (2005) Studies on the ecology and conservation of butterflies in Europe. Vol 2: species ecology along a European gradient: Maculinea butterflies as a model. Pensoft Publishers, Sofia, Moscow
Settele J, Shreeve T, Konvička M, Van Dyck H (2009) Ecology of butterflies in Europe. Cambridge University Press, Cambridge
Sielezniew M, Stankiewicz AM (2008) Myrmica sabuleti (Hymenoptera: Formicidae) not necessary for the survival of the population of Phengaris (Maculinea) arion (Lepidoptera: Lycaenidae) in eastern Poland: lower host-ant specificity or evidence for geographical variation of an endangered social parasite? Eur J Entomol 105:637–641
Sielezniew M, Buszko J, Stankiewicz AM (2005) Maculinea arion in Poland: distribution, ecology and prospects of conservation. In: Settele J, Kühn E, Thomas JA (eds) Studies on the ecology and conservation of butterflies in Europe, Vol 2: species ecology along a European gradient: Maculinea butterflies as a model. Pensoft Publishers, Sofia, Moscow, pp 231–233
Sielezniew M, Dziekańska I, Stankiewicz-Fiedurek AM (2010a) Multiple host-ant use by the predatory social parasite Phengaris (=Maculinea) arion (Lepidoptera, Lycaenidae). J Insect Conserv 14:141–149
Sielezniew M, Włostowski M, Dziekańska I (2010b) Myrmica schencki (Hymenoptera: Formicidae) as the primary host of Phengaris (Maculinea) arion (Lepidoptera: Lycaenidae) at heathlands in eastern Poland. Sociobiology 55:95–106
Sielezniew M, Patricelli D, Dziekańska I, Barbero F, Bonelli S, Casacci LP, Witek M, Balletto E (2010c) The first record of Myrmica lonae (Hymenoptera: Formicidae) as a host of the socially parasitic large blue butterfly Phengaris (Maculinea) arion (Lepidoptera: Lycaenidae). Sociobiology 56:465–475
Sigaard P, Pertoldi C, Madsen AB, Søgaard B, Loeschcke V (2008) Patterns of genetic variation in isolated Danish populations of the endangered butterfly Euphydryas aurinia. Biol J Linn Soc 95:677–687
Spitzer L, Dandová J, Jašková V, Beneš J, Konvička M (2009) The large blue butterfly, Phengaris [Maculinea] arion, as a conservation umbrella on a landscape scale: the case of the Czech Carpathians. Ecol Indic 9:1056–1063
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thissen D, Steinberg L, Kuang D (2002) Quick and easy Implementation of the Benjamini-Hochberg procedure for controlling the false positive rate in multiple comparisons. J Educ Behav Stat 27:77–83
Thomas JA (1995) The ecology and conservation of Maculinea arion and other European species of large blue butterfly. In: Pullin AS (ed) Ecology and conservation of butterflies. Chapman and Hall, London, pp 180–197
Thomas JA (2002) Larval niche selection and evening exposure enhance adoption of a predacious social parasite, Maculinea arion (large blue butterfly), by Myrmica. Oecologia 132:531–537
Thomas JA, Settele J (2004) Butterfly mimics of ants. Nature 432:283–284
Thomas JA, Elmes GW, Wardlaw JC, Woyciechowski M (1989) Host specificity among Maculinea butterflies in Myrmica ant nests. Oecologia 79:425–457
Thomas JA, Simcox DJ, Wardlaw JC, Elmes GW, Hochberg ME, Clarke RT (1998) Effects of latitude, altitude and climate on the habitat and conservation of the endangered butterfly Maculinea arion and its Myrmica ant hosts. J Insect Conserv 2:39–46
Thomas JA, Simcox DJ, Clarke RT (2009) Successful conservation of a threatened Maculinea butterfly. Science 325:80–83
van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Van Swaay CAM, Warren MS (1999) Red data book of European butterflies (Rhopalocera). Nature and Environment, No. 99. Council of Europe Publishing, Strasbourg
Van Swaay C, Cuttelod A, Collins S, Maes D, Lopez Munguira M, Šašić M, Settele J, Verovnik R, Verstrael T, Warren M, Wiemers M, Wynhof I (2010) European red list of butterflies. Publications Office of the European Union, Luxembourg
Vandewoestijne S, Schtickzelle N, Baguette M (2008) Positive correlation between genetic diversity and fitness in a large, well-connected metapopulation. BMC Biology 6:46
Vila M, Cassel Lundhagen A, Thuman KA, Stone JR, Björklund M (2006) A new conservation unit in the butterfly Erebia triaria (Nymphalidae) as revealed by nuclear and mitochondrial markers. Ann Zool Fenn 43:72–79
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wynne IR, Loxdale HD, Brookes CP, Woiwod IP (2003) Genetic structure of fragmented November moth (Lepidoptera: Geometridae) populations in farmland. Biol J Linn Soc 78:467–477
Zeisset I, Als TD, Settele J, Boomsma JJ (2005) Microsatellite markers for the large blue butterflies Maculinea nausithous and Maculinea alcon (Lepidoptera: Lycaenidae) and their amplification in other Maculinea species. Mol Ecol Notes 5:165–166
Acknowledgments
We are indebted to Paweł Borkowski, Cezary Bystrowski, Arkadiusz Dębała, Izabela Dziekańska, Krzysztof Frąckiel, Adam Górnicki, Mariusz Gwardian, Tadeusz Janik, Przemysław Klimczuk and Krzysztof Pałka for the precise location of sites of P. arion and/or for logistic help during field studies. Irma Wynhoff and a few anonymous reviewers made valuable comments on the earlier drafts of the manuscript. The Polish Minister of the Environment issued the relevant permissions for our studies. This work was supported by the Polish Ministry of Science and Higher Education (grant no 2 P04G 024 30).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Sielezniew, M., Rutkowski, R. Population isolation rather than ecological variation explains the genetic structure of endangered myrmecophilous butterfly Phengaris (=Maculinea) arion . J Insect Conserv 16, 39–50 (2012). https://doi.org/10.1007/s10841-011-9392-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10841-011-9392-9