Abstract
Epigenetic processes including DNA methylation play a pivotal role in regulating the genes that control plant development. In contrast to in planta development, the contribution of DNA methylation to the morphogenic processes that are induced in vitro are much less recognised. Hence, in the present study, we analysed the impact of DNA methylation on somatic embryogenesis (SE) that was induced in Arabidopsis. The results demonstrated a decrease in the global DNA methylation level during SE that contrasted with the up-regulation of MET1 and CMT3 DNA methylases and the down-regulation of DNA demethylases (ROS1, DME and DML2). Hence, the global DNA methylation level appears not to correlate with the transcriptional activity of the genes encoding DNA methylases/demethylases, thereby implying the complexity of the regulatory mechanism that controls the DNA methylation status of the SE-epigenome. Moreover, distinct changes in the expression level of the SE-regulatory genes were indicated in the 5-AzaC-treated and DNA methylase mutant cultures. Accordingly, a significant repression of the LEC2, LEC1 and BBM genes was found in the 5-AzaC-treated culture that was incapable of SE induction. In contrast, the distinct up-regulation of these genes was observed in the drm1drm2 and drm1drm2cmt3 mutant cultures with an improved embryogenic response. The modulated expression of DNA methylase genes and the significantly modified embryogenic response of the met1 and drm mutants imply that both the maintenance and the de novo pathway of DNA methylation are engaged in the regulation of SE in Arabidopsis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Somatic embryogenesis (SE) is a plant-specific developmental process that involves the induction of the embryogenic programme in somatic cells, which results in the formation of somatic embryos that are capable of regenerating complete plants. The transition of already differentiated cells into embryogenic ones requires extensive changes in the somatic cell transcriptome. Accordingly, rapid changes in the gene expression patterns that accompany SE induction have been reported in various plants (Elhiti et al. 2013) including Arabidopsis (Gliwicka et al. 2013; Wickramasuriya and Dunwell 2015). The reprogramming of the cell transcriptome that results in the release of a new developmental programme is associated with extensive changes in the chromatin structure, which involves chemical modifications of DNA and histones (He et al. 2011a; De-la-Peña et al. 2015; Mozgová et al. 2017). Among the epigenetic modifications that control gene expression, the methylation of DNA is considered to play a pivotal role in plant development (Zhang et al. 2010; Turck and Coupland 2014; Victoria et al. 2018).
The methylation of plant DNA involves the addition of a methyl group to the carbon-5 of cytosine at the CpG, CpNpG and CpNpN (where N could be any nucleotide except G) sequences in DNA, which results in an increase of the content of 5-methyl cytosine (5 mC) in the genomic DNA (Law and Jacobsen 2010). Three types of DNA methylases have been found in plants including METHYLTRANSFERASE 1 (MET1), CHROMOMETHYLASE 3 (CMT3) and DOMAINS REARRANGED METHYLTRANSFERASE (DRM1 and DRM2) (Lindroth et al. 2001; Cao and Jacobsen 2002a; Zhang et al. 2006). In Arabidopsis thaliana, DRMs are required for de novo methylation while MET1 and CMT3 maintain the methylation pattern during DNA replication (Zhang et al. 2010).
DNA methylases cooperate with other proteins and the recruitment of MET1 to DNA requires the activity of the variant in methylation (VIM/ORTHRUS) proteins that recognise the hemimethylated CpG sites that are generated during DNA replication (Shook and Richards 2014). The accession of DNA methyltransferases to DNA is supported by DECREASED IN DNA METHYLATION (DDM1), which is a SWI/SNF chromatin-remodeling factor (Jeddeloh et al. 1999).
In controlling genome-wide DNA methylation patterns, DNA methylation is accompanied by active DNA demethylation that is carried out by the DNA glycosylase family of DNA demethylases in plants (Penterman et al. 2007; Stroud et al. 2013). The Arabidopsis genome encodes four DNA demethylases, including DEMETER (DME), REPRESSOR OF SILENCING 1 (ROS1)/DEMETER-LIKE 1 (DML1), DML2 and DML3. ROS1 is a major DNA demethylase that is involved in the dynamic transcriptional regulation of the genome (Gong et al. 2002) that plays a role in the developmental processes and biotic and abiotic stress responses (Yamamuro et al. 2014; Schumann et al. 2017). DME is required for the expression of specific imprinted maternal alleles during seed development, while the biological function of DML2 and DML3 as yet remains mostly unknown (Bauer and Fischer 2011).
DNA methylation has been recognised as a mechanism that suppresses gene expression because the accumulation of 5 mC has been observed in the heterochromatin regions, imprinted genes, repetitive sequences and transposons (Tariq and Paszkowski 2004; Teixeira and Colot 2010). However, in a large number of expressed genes, the promoters and the transcribed regions of the genes (‘gene body’) are methylated (He et al. 2011b; Jones 2012). The frequent methylation of the gene body of constitutively expressed housekeeping genes suggests a homeostatic function of this type of methylation (Zilberman 2017). In addition, methylation of the gene body might also be related to the regulation of the responses of genes to internal or external cues (Aceituno et al. 2008). Thus, the function of CG methylation within the transcribed regions of genes is currently unclear (Bewick and Schmitz 2017).
During the life cycle of Arabidopsis, the global level of DNA methylation undergoes dynamic changes and the content of 5 mC decreases in gametogenesis and increases after fertilisation and during embryo development (Jullien et al. 2012; Bouyer et al. 2017). In adult plants, variations in the level and pattern of methylation have been observed between the plant organs but the overall trend is that the 5 mC content increases during the aging and maturation of a plant (Ruiz-García et al. 2005; Widman et al. 2014).
A nucleotide analogue, 5-Azacitidine (5-AzaC), of the DNA demethylation activity is commonly applied to study the role of DNA methylation in the developmental processes. 5-AzaC is randomly incorporated into a newly synthesised DNA strand instead of cytosine and, as a result, a dose- and time-dependent decrease in the DNA methyltransferase activity can be observed that is followed by genome hypomethylation at random sequences (Christman 2002; Issa and Kantarjian 2009). Treatment with 5-AzaC has been shown to induce various plant phenotypes including dwarfism, early flowering and an inhibition of vegetative growth (Kondo et al. 2006). Although 5-AzaC treatment has also been indicated to impact the morphogenic processes that are induced in vitro including the capacity of a tissue for SE (Santos and Fevereiro 2002; Yamamoto et al. 2005; Tokuji et al. 2011; Fraga et al. 2012; Teyssier et al. 2014; Solís et al. 2015), our knowledge about the role of this modification in the epigenetic regulation of the genes that control the developmental plasticity of somatic plant cells is still limited.
Hence, we were motivated to study the function of DNA methylation in regulating SE in Arabidopsis, which is a model plant that offers a rapid and efficient culture system to identify the molecular determinants that are involved in the embryogenic transition induced in somatic cells (Gaj 2001; Wójcikowska and Gaj 2016). Different analytical approaches that aimed at revealing (1) the impact of both 5-AzaC-treatment and the mutations that affect the DNA methylase genes (drm1drm2 and drm1drm2cmt3) on the embryogenic capacity of a tissue and the expression profiles of the LEC1, LEC2 and BBM genes that play a key role in SE induction (2) changes in the global DNA methylation level during SE and (3) the contribution of the genes encoding different DNA methylases to SE induction. The results indicated that the DNA methylation status of the explants impacts both the capacity of a culture for SE and the expression level of the SE-involved genes. The results of the DNA methylase gene profiling together with an analysis of the mutants that were defective in DNA methylases suggest that both the maintenance and the de novo pathway of DNA methylation are engaged in the regulation of SE in Arabidopsis.
Materials and methods
Plant material
Arabidopsis thaliana (L.) Heynh. plants of the Columbia (Col-0) WT ecotype and the insertional mutant lines in the Col-0 background were used in the study (Table S1). The seeds were purchased from NASC (The Nottingham Arabidopsis Stock Centre, UK; http://arabidopsis.info/). Selection of homozygous mutants was conducted following the NASC standard protocol (http://signal.salk.edu/tdnaprimers.2.html).
In vitro culture of the explants
In order to induce SE, immature zygotic embryos (IZEs) at the stage of green, fully developed cotyledons were cultured in vitro following a standard protocol (Gaj 2001). The standard medium used for SE induction (E5) contained a basal B5 medium (Gamborg et al. 1968) that was supplemented with 20 g L−1 sucrose, agar (8 g L−1) and 5 μM of 2,4-dichlorophenoxyacetic acid (2,4-D; Sigma-Aldrich). In the control culture, the explants were induced on an E5 medium without 2,4-D (E0), which resulted in seedling development. In some experiments, the E5 medium was supplemented with 10 μM of 5-AzaC (5-Azacitidine; Sigma-Aldrich).
Ten explants were cultured in one Petri dish and 30 explants in three replicates from each culture combination were analysed. The capacity for SE was evaluated in three-week-old cultures. Two parameters of embryogenic potential were evaluated: SE efficiency—the frequency of the explants that produced somatic embryos and SE productivity—the average number of somatic embryos that developed per embryogenic explant.
Explants that were cultured for 0 d; 3 d; 5 d; 10 d; 15 d (d = day of culture) on the E5 or E0 medium were collected to isolate RNA and DNA. A small fraction (< 10%) of the explants that were cultured on E5 failed to induce SE and developed a non-embryogenic callus that was also taken for analysis (15 days). All of the molecular analyses of the SE culture including RT-qPCR and ELISA were conducted in three biological and two technical replicates.
Plant growth and in vitro culture conditions
The plants that were used as the source of the IZE explants were grown in Jiffy-7 peat pots (Jiffy) in a ‘walk-in’ type phytotron under controlled conditions at 22 °C under a 16 h photoperiod of 100 µM m−2 s−1 white, fluorescent light. The plant materials that were grown in sterile conditions were kept at 23 °C under a 16 h photoperiod of 40 µM m−2 s−1 white, fluorescent light.
Total RNA isolation and RT-qPCR
An RNAqueous Kit (Ambion) was used to isolate total RNA. The concentration and purity of RNA was evaluated using an ND-1000 spectrophotometer (NanoDrop). In order to prevent DNA contamination, the RNA was treated with RQ1 RNase-free DNase I (Promega) following the manufacturer’s instructions. First strand cDNA was produced in a 20 µL reaction volume using a RevertAid First Strand cDNA Synthesis Kit (Fermentas).
The product of the reverse transcription was used to evaluate the expression level of the selected genes (Table S2). RT-qPCR was carried using a LightCycler® 480 SYBR Green I Master (Roche), the appropriate mix of the master mix, cDNA and water were used for the RT-qPCR reactions (Table S3). A LightCycler 480 (Roche) real-time detection system was used under the following reaction conditions: denaturation one repeat of 10 min at 95 °C followed by 45 repeats of 10 s at 95 °C, 8 s at 55 °C, 12 s at 72 °C and 5 s at 80 °C. Denaturation for the melt curve analysis was conducted at 95 °C followed by 15 s at 65 °C and then heating to 95 °C (0.1 °C/s, with continuous fluorescence measurement).
Primary data analysis was performed using LightCycler Software 4.0 (Roche). The relative RNA levels were calculated and normalised to an internal control—the At4g27090 gene encoded 60S ribosomal protein (Thellin et al. 1999). In all of the analysed tissue samples, the control gene displayed a constant expression pattern with Cp = 18 ± 1 (Figs. 2, 3 and 4b, c) and Cp = 23 ± 1 (Figs. 4a and S1).
DNA isolation and DNA methylation analysis with ELISA
The modified micro C-TAB method was used to extract genomic DNA from the explants that had been cultured on the E5 medium for 0, 3, 5, 10 and 15 days (Doyle and Doyle 1987). The global DNA methylation level was evaluated using spectrophotometric methods—a 5-mC DNA ELISA kit (ZymoResearch) following the manufacturer’s protocol. The light absorbance was analysed on a Victor X5 multilabel reader system (PerkinElmer).
Statistical analyses
The Student’s t test was used to calculate any significant differences (at P = 0.05) between the combinations that were being compared.
Results
Reduced embryogenic response of the explants treated with 5-AzaC
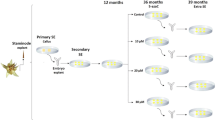
The embryogenic capacity of the Col-0 explants that were treated with 5-AzaC was studied. In the control culture that was induced on the E5 medium, the explants formed somatic embryos rapidly and efficiently while on the medium that had been supplemented with 10 µM of 5-AzaC, the SE response was drastically inhibited and the explants massively produced white, non-embryogenic callus tissue (Fig. 1). Treating the explants with 5-AzaC reduced both the efficiency and productivity of SE and as a result only 5% of the explants were able to undergo SE induction and the average number of somatic embryos produced by an embryogenic explant was decreased by more than half compared to the control culture. Since no signs of tissue lethality were observed in the treated cultures, we hypothesised that the inhibition of SE was not a result of the toxic effect of 5-AzaC on cell metabolism but that it resulted from DNA hypomethylation-associated effects including changes in the gene transcription.
The expression profiles of the SE-involved TF genes in response to 5-AzaC treatment
RT-qPCR analysis was used to analyse the expression of the genes encoding the transcription factors (TFs) of the documented regulatory function in SE induction in Arabidopsis including LEC1 (LEAFY COTYLEDON1), LEC2 (LEAFY COTYLEDON2) and BBM (BBM BABY BOOM). The gene expression profiles in response to 5-AzaC treatment of the WT (Col-0) culture were evaluated.
The results showed that 5-AzaC treatment caused significant changes in the transcription profiles of the genes that were analysed (Fig. 2). Under 5-AzaC treatment, the strongest inhibition of the transcript level was observed for BBM and LEC2 whose expression was highly repressed up to 32 (LEC2) and 11 (BBM)—fold in comparison to transcript level observed in control E5 culture after the transient up-regulation in the early culture (5 days). In contrast, the LEC1 expression profile in the culture that was induced on the E5 + 5-AzaC medium was similar to the one that was observed in the control culture (E5) and an increased transcript accumulation was indicated at the early (5 days) and advanced (15 days) SE stages.
Expression profiles of SE-involved TF genes in the Col-0 (WT) embryogenic culture induced on E5 (control) and E5 + 10 µM 5-AzaC medium. Relative transcript level was normalised to the internal control (At4g27090) and calibrated to the 0 day of culture. Values significantly different to 0 day are indicated with an asterisk; Values significantly different to control (E5) at corresponding day of culture are indicated with a hashtag (Student’s t test, P < 0.05). Error bars indicate the standard deviation (SD)
The expression profiles of the SE-involved TF genes in the DNA methylation-related mutants
To further assess the impact of DNA methylation on SE induction, we profiled the expression of the LEC1, LEC2 and BBM genes in the SE-induced explants of the drm1drm2 and drm1drm2cmt3 mutants (henceforth referred to as dd and ddc). The results demonstrated that the expression profiles of LEC1, LEC2 and BBM genes were similar in the dd and ddc mutant cultures and that the analysed genes showed a significantly increased expression in the mutant explants that were induced on the E5 medium (Fig. 3). The genes that were analysed differed in the relative level of gene transcripts and LEC1 displayed the highest (up to 77-fold) and BBM the lowest (up to threefold) stimulation of gene expression in the mutant cultures. In addition, we observed that an increase in the gene transcript level was higher in the mutant (Fig. 3) than in WT (Fig. 2) culture that was induced on E5 for all of the SE-regulatory genes that were analysed.
Expression profiles of LEC1, LEC2 and BBM genes that have a regulatory role in SE induction in the drm1drm2 (dd) and drm1drm2cmt3 (ddc) mutant cultures induced on E5 medium. The relative transcript level was normalised to the internal control (At4g27090) and calibrated to the 0 day culture. The values that were significantly different to 0 day are indicated with an asterisk (Student’s t test, P < 0.05). Error bars indicate the standard deviation (SD)
Expression of the genes that are involved in DNA methylation and DNA demethylation during SE
To further explore the DNA methylation-related processes that are involved in the molecular mechanism that governs the embryogenic response in plant somatic cells, we evaluated the expression profiles of the genes encoding DNA methylases (MET1, CMT3, DRM1 and DRM2) in the WT (Col-0) explants that had been cultured on the E5 (auxin) vs the E0 (auxin-free) medium. In contrast to the SE-promoting E5 medium, the explants that had been cultured on E0 were not able induce SE and they developed into seedlings instead.
Although we found that the DNA methylase genes were expressed in both of the culture conditions that were analysed, distinct differences in the gene expression profiles and transcript accumulation between the genes and the culture conditions were observed (Fig. 4). Accordingly, a high accumulation of MET1 and CMT3 in the SE culture (3–10 days) was observed that was specific to the E5 medium (Fig. 4a). Compared to MET1 and CMT3, the genes that control de novo DNA methylation, DRM1 and DRM2, displayed a significantly lower expression level during SE and their transcription profiles on E5 and E0 were similar. A distinct difference between the expression profiles of DRM1 and DRM2 genes in the advanced stage of SE culture was observed and at 10–15 days of the culture, a down-regulated DRM1 transcription contrasted with an up-regulated expression of DRM2. Moreover, we found that the accumulation of the DRM2 transcripts in the SE culture significantly exceeded (up 4000-fold) the transcript level of the DRM1 gene (Fig. S1).
Expression level of the genes encoding DNA methylases, MET1, CMT3, DRM1, DRM2 (a); DNA-methylation related proteins, DDM1, VIM1 (b) and the DNA demethylases, ROS1, DME, DML2 (c) in the Col-0 explants cultured on the E5 and the E0 medium. Relative transcript level was normalised to the internal control (At4g27090) and calibrated to the 0 day of culture. Values significantly different from 0 day are indicated with an asterisk; Values significantly different from E0 are indicated with a hashtag (Student’s t test, P < 0.05). Error bars indicate the standard deviation (SD)
We also investigated the expression of the DDM1 and VIM1 genes encoding the proteins that are related to DNA methylation. We found that the transcription of DDM1 and VIM1 was significantly modulated during SE and that their expression patterns were similar (Fig. 4b). Accordingly, a substantial up-regulation of the DDM1 and VIM1 transcripts was observed during SE induction. In contrast to the genes encoding the DNA methylases, the DNA methylation-related genes, DDM1 and VIM1 displayed a down-regulation during SE (E5) and an up-regulation in the explants that developed seedlings on the E0 medium.
In addition to the genes that are involved in DNA methylation, we also analysed the expression level of three demethylase genes, including (ROS1) REPRESSOR OF SILENCING 1, (DME) DEMETER and (DML2) DEMETER LIKE 2 in the explants that had been cultured on E5 and E0 (Fig. 4c). We found that in the SE culture that had been induced on the E5 medium, all of the analysed DNA demethylase genes were distinctly down-regulated up to 23–70% of the level that was found in the freshly isolated (0 day) explants. Similarly, the DML2 expression was distinctly reduced in the seedlings that were developing on E0 while the transcription of the other two analysed genes, DME and ROS1, was not affected on E0, except for a transient up-regulation of the ROS1 at 5 days.
Embryogenic capacity of the DNA methylation-related mutants
In addition to the analysis of the gene expression, we evaluated the capacity for SE of the insertional mutants that were affected in the genes encoding the DNA methylases (met1, cmt3, dd and ddc) and proteins that are involved in DNA methylation (ddm1 and vim1) (Fig. 5). The results showed a distinctly decreased SE response of the met1 and two other mutants, ddm1 and vim1, with an insert in the DNA methylation-related genes. In contrast, cultures with mutations in the DRM1 and DRM2 methylase genes (dd and ddc) were found to display the opposite phenotype in the culture on the E5 medium and they showed an increased capacity for somatic embryo formation.
Efficiency and productivity of the SE cultures derived from the mutants that were defective in the genes encoding DNA methylases (met1, cmt3, drm1drm2, drm1drm2cmt3) and the proteins involved in DNA methylation (ddm1 and vim1). Values significantly different from the control culture (Col-0) are indicated with an asterisk (Student’s t test, P < 0.05). Error bars indicate the standard deviation (SD)
Global DNA methylation level in SE
Considering that the genes that are involved in DNA methylation displayed different expressions in the embryogenic culture, we expected that the level of DNA methylation would be modulated during SE. Therefore, we investigated the global level of 5 mC in the DNA in the explants at various time points (0, 3, 5, 10, 15 days) of the SE culture. In addition to the embryogenic explants, the non-embryogenic callus tissue that had been developed by a small fraction (less than 10%) of the E5-cultured explants was analysed.
The results indicated a significant and progressive decrease in the global level of DNA methylation in the SE-induced explants. Accordingly, the level of 5 mC was up to 30% lower (15 days) in the SE culture than in the freshly isolated 0 day explants (Fig. 6). We found that both the embryogenic (SE) and non-embryogenic (callus) tissue showed a similar level of the global DNA methylation.
Global changes in the DNA methylation level in the Col-0 explants induced towards SE on the E5 medium (0, 3, 5, 10 and 15 days) and in the non-embryogenic callus. Values significantly different from 0 day are indicated with an asterisk (Student’s t test, P < 0.05). Error bars indicate the standard deviation (SD).4
Discussion
Recently, significant progress has been made in identifying genes and revealing their biological functions in the genetic mechanisms that control the embryogenic transition in somatic cells using the SE culture of Arabidopsis (reviewed in Nowak and Gaj 2016). In contrast, our understanding of the epigenetic processes that contribute to the regulation of SE remains very limited. Hence, the present work was focused on an analysis of the role of DNA methylation in SE induction in Arabidopsis explants that were cultured in vitro.
5-AzaC treatment and DNA methylase-related mutations affect both the expression of the SE-involved TF genes and the embryogenic response
5-AzaC treatment was indicated as distinctly changing the gene expression in the plants (Chang and Pikaard 2005; Song et al. 2017) and in vitro cultured plant cells/tissue and both the stimulation and repression of the gene transcription has been reported (Berdasco et al. 2008; Tokuji et al. 2011; Nic-Can et al. 2013). Consistent with the diverse effects of 5-AzaC on gene expression level, the hypo- and hypermethylation of DNA in response to this chemical were reported in an embryogenic culture of Acca sellowiana, which were dependent on the plant genotype and the supplementation of the culture medium with 2,4-D (Fraga et al. 2012).
In the present study, we demonstrated that in Arabidopsis, similar to the embryogenic cultures of other plants (Santos and Fevereiro 2002; Yamamoto et al. 2005; Teyssier et al. 2014), treating the tissue with 5-AzaC severely and negatively affected the embryogenic response of the explants. We observed that the decreased embryogenic response of the 5-AzaC-treated explants is accompanied by a deregulated expression of LEC1, LEC2 and BBM that have a key regulatory role in SE induction. Similarly, 5-AzaC distinctly reduced the expression level of the shoot regeneration-related TF genes in the callus of Arabidopsis (Tokuji et al. 2011). LEC1 and LEC2, which are the master regulators of zygotic embryogenesis in Arabidopsis (Braybrook and Harada 2008), were found to be essential for SE induction (Gaj et al. 2005). LEC1 and LEC2 contribute to SE induction via the regulation of the auxin response genes (Braybrook et al. 2006) and LEC2 was indicated as activating auxin biosynthesis during SE (Wójcikowska et al. 2013). Recently, the LEC1, LEC2 and BBM genes were indicated as functioning in the same molecular pathway in which BBM transcriptionally regulates LEC1 and LEC2 to induce SE (Horstman et al. 2017). Relevant to the BBM-mediated SE induction mechanism, we found the BBM expression to be distinctly repressed in the 5-AzaC-treated culture that was incapable of SE induction. 5-AzaC was also found to strongly inhibit the embryogenic response in coffee by decreasing the expression of LEC1 and BBM1 (Nic-Can et al. 2013). Given that 5-AzaC-treatment has been reported to distinctly reduce the global methylation level in the in vitro cultured tissue of different plants that have been cultured in vitro, including Arabidopsis (Tokuji et al. 2011), rape and barley (Solís et al. 2015) and coffee (Nic-Can et al. 2013), we assumed that the repression of the SE-regulatory genes that are associated with a decrease in the SE response is related to the demethylation of DNA.
In contrast to the 5-AzaC treated cultures, in the dd and ddc mutant cultures of the increased embryogenic response expression of BBM, LEC1 and LEC2 was significantly up-regulated (Fig. 3). Similarly, the significant up-regulation of the genes that are located in the euchromatin was reported in an ddc mutant (Zhang et al. 2006) and the induction of the dd seedling explants on the auxin medium resulted in a continuous increase of the BBM expression that was followed by accelerated callus production (Jiang et al. 2015). Relevant to the up-regulated expression of LEC1, LEC2 and BBM in the dd/ddc mutant cultures with an increased SE response, the overexpression of these TFs was demonstrated to be sufficient to induce SE in Arabidopsis (Lotan et al. 1998; Stone et al. 2001; Boutilier et al. 2002) and other plants (Heidmann et al. 2011; Guo et al. 2013). Collectively, the contrasting expression level of the LEC1, LEC2 and BBM genes, in particular BBM, a positive regulator of LEC1 and LEC2 during SE induction (Horstman et al. 2017), which was found in the dd/ddc versus 5-AzaC-treated cultures might account for the opposite embryogenic capacity of these cultures.
The oppose expression profiles of the SE-regulatory genes that we found in the 5-AzaC-treated and dd/ddc mutant cultures might be related to the different impact of the 5-AzaC and dd/ddm mutations on the cell methylome. By trapping the DNA methyltransferase in the replication fork, 5-AzaC causes passive DNA demethylation that results in a global and stochastic reduction of the 5metC content in the genome (Chang and Pikaard 2005; Szyf 2009). In contrast to 5-AzaC, the mutations in CMT3 and DRM methylases only cause a mild decrease in the overall DNA methylation level (Zhang et al. 2006; Table S4) and the demethylation is specific to the DNA sequences (Cokus et al. 2008; Stroud et al. 2013, 2014).
In summary, these results infer a role of DNA methylation in the epigenetic control of the embryogenic response in Arabidopsis and suggest that DNA methylation might contribute to SE induction by impacting the expression of the key regulators of SE.
SE induction is associated with the decreased level of global DNA methylation
It is believed that the demethylation of DNA is characteristic of the open-chromatin state that is required to redirect the somatic cells into embryogenic development (Grafi et al. 2011; Miguel and Marum 2011). In line with this postulate, we observed a significantly decreased level of 5 mC in the embryogenic culture of Arabidopsis and the reduction in the DNA methylation level has also been associated with SE induction in other plants (Santos and Fevereiro 2002; Xu et al. 2004; Noceda et al. 2009; Rodríguez-Sanz et al. 2014; Teyssier et al. 2014). However, in some plants, an increased (Fraga et al. 2012; Rival et al. 2013), differentially modulated (Leljak-Levanić et al. 2004; Nic-Can et al. 2013) and steady (Parra et al. 2001) level of DNA methylation during SE has been reported. The diversity in the SE-associated DNA methylation levels suggests that various exo- and endogenous culture factors rather than the induction of embryonic development per se appear to contribute to the global DNA methylation level in cultured tissue (Huang et al. 2012; Rival et al. 2013; Li et al. 2014; Rathore et al. 2014). In support of this assumption, we found that the level of 5 mC in the embryogenic culture was similar to the one that was observed in the non-embryogenic callus that was formed by the explants that failed in the embryogenic transition.
In conclusion, the global content of 5 mC in the cultured tissue does not appear to be specific to the type of morphogenic development that is induced but rather reflects unspecific epigenetic status of the explant cells and their dedifferentiation in response to the in vitro culture conditions. Thus, in order to reveal the impact of DNA methylation on SE regulation, the gene-specific pattern of DNA methylation needs to be revealed.
Both the maintenance and de novo pathways of DNA methylation are involved in SE induction
The role of DNA methylation in the embryogenic response of the Arabidopsis explants was further inferred by the significantly modulated expression of the genes encoding DNA methylases during SE. We found that the genes of the methylases that are involved in the maintenance (MET1, CMT3) and de novo (DRM1 and DRM2) DNA methylation are expressed differently during SE. We found the significant up-regulation of the MET1 and CMT3 genes during SE in Arabidopsis and, consistent with this result, a higher activity of these genes has been reported in zygotic embryos, in particular during the early stages of embryo formation, compared to other organs of Arabidopsis (Xiang et al. 2011; Jullien et al. 2012; Ashapkin et al. 2016). Thus, it appears that the enhanced activity of the MET1 and CMT3 genes is generally associated with embryonic development and might reflect a requirement for the propagation of the DNA methylation pattern in the intensively dividing pro-embryonic and embryonic cells. In support of this assumption, the period with the highest expression of the MET1 and CMT3 genes coincides with the time of the intensive cell divisions that are associated with the acquisition of embryonic fate by the explant cells (Kurczyńska et al. 2007).
Expression profiling of the DNA methylase genes on the E5 versus the E0 medium infers that auxin treatment activates MET1 and CMT3 transcription. A positive impact of auxin on the CMT3 expression has also been reported by others (Parizot et al. 2010; Shemer et al. 2015). In support of the auxin-controlled expression of CMT3 is the presence of the auxin-responsive motif, AuxRE, in the promoter of this gene (http://arabidopsis.med.ohio-state.edu/AtcisDB/), which implies that ARFs, the core elements of auxin signalling that are believed to play roles in SE induction in Arabidopsis (Weijers and Wagner 2016; Wójcikowska and Gaj 2017), might regulate CMT3 in response to auxin treatment. In contrast to CMT3, the impact of auxin on MET1 up-regulation in SE appears to be indirect due to the lack of auxin-responsive motifs in the gene promoter.
The impaired embryogenic capacity of the ddm1 and vim1 mutants (present results) suggests that similar to in planta development, the DNA methylation-related proteins VIM1 and DDM1 support the function of MET1 methylases during SE (Zemach et al. 2013; Shook and Richards 2014). The VIM1 protein contributes to the recruitment of MET1 to a newly replicated DNA strand (Shook and Richards 2014). Consistent with this, we found that the up-regulation of VIM1 was associated with an increased MET1 transcription in SE.
We observed that the explants of the cmt3 mutant were unaffected in SE response and in contrast, the met1 mutant was significantly defective in SE induction. The contrasting embryogenic capacity of the cmt3 and met1 mutants suggests that the contribution of the CMT3 and MET1 methylases to SE appears to be different. The role of CMT3 in the callus-mediated shoot regeneration of Arabidopsis (Shemer et al. 2015) might suggest that in the SE system of Arabidopsis in which marginal callus production accompanies the direct development of somatic embryos from explant cells (Gaj 2001; Kurczyńska et al. 2007), the function of CMT3 might be not critical for SE induction. In contrast, the importance of MET1 activity for SE induction might be related with the role of the enzyme in the hormone-related processes that play a central role in SE induction (Nic-Can and Loyola-Vargas 2016). Accordingly, MET1 has been indicated as impacting the expression of ARF3 and WUS, which are involved in hormone signalling (Li et al. 2011), and the PIN1 gene that is engaged in auxin polar transport in ZE (Xiao et al. 2006).
In the present study we found that an increased expression level of the CMT3 and MET1 DNA methylases contrasts with a decreased global level of DNA methylation in the embryogenic culture. Since, DNA methylation is accompanied by active DNA demethylation and the balance between these antagonistic processes is believed to control the pattern of DNA methylation in plant development including the early stages of embryogenesis (Penterman et al. 2007; Lei et al. 2015; Bouyer et al. 2017), we also analysed the expression of the genes encoding DNA demethylases during SE induction and a distinct down-regulation of ROS1, DME and DML2 genes in the SE-induced explants was observed. Thus, the reduction of the global DNA methylation level that was found in the embryogenic culture (present results) appears not to directly result from the transcriptional activity of the genes encoding DNA methylases and demethylases. In support of this assumption, an analysis of the rice genome showed that the level of cytosine methylation is not directly correlated with the activity of the DNA methylases (Teerawanichpan et al. 2009). In addition, an analysis of the triple mutant that was affected in ROS1, DML2 and DML3 genes indicated an unchanged DNA methylation level although discrete loci were hypermethylated (Penterman et al. 2007). It was postulated that the lack of a direct correlation between the DNA methylase activity and the 5mC level might be a consequence of the complexity of the interactions that control the balance between DNA replication, the de novo maintenance DNA methylation and demethylation (Hsieh et al. 2009). The complex interplay between DNA methylation and demethylation exemplifies the fine tuning of the expression of ROS1 to variations in the DNA methylation level in which the methylation and demethylation of the gene promoter region promote and suppress gene transcription, respectively (Lei et al. 2015). Relevant to this model, an extremely low level of ROS1 transcripts was indicated in mutants that were impaired in DNA methylation, including met1, which had a significantly reduced overall level of DNA methylation (Cokus et al. 2008; Williams et al. 2015).
Our analyses indicate that in the SE of Arabidopsis de novo DNA methylation appears to mainly be controlled by DRM2 methylase due to the high expression of DRM2 at all of the monitored time points of the culture (Fig. S1). A distinctly higher level of DRM2 than DRM1 transcripts was also shown to be characteristic of in planta development including zygotic embryo formation in Arabidopsis (Xiang et al. 2011; Jullien et al. 2012; Ashapkin et al. 2016). Moreover, we found that the auxin medium positively impacted the expression of the DRM genes as was reported in a callus culture induced from Arabidopsis seedlings (Jiang et al. 2015). The role of de novo DNA methylation in the control of SE induction also infers the finding that the distinct up-regulation of the SE-regulators (LEC1, LEC2 and BBM) in the dd and ddc mutant cultures is associated with improved SE response (present results). The reduced content of non-CG methylated sites in DNA causing chromatin de-condensation and de-repression of genes might account for the increased LEC1, LEC2 and BBM expression that was indicated in the culture of DRM-defected mutants (Cao and Jacobsen 2002b; Henderson and Jacobsen 2008).
Conclusions
Modification of the DNA methylation state of the cultured explants via 5-AzaC treatment and mutations in the genes encoding DNA methylases (dd/ddc) distinctly alters both the embryogenic response and the expression profiles of LEC2, LEC1, BBM genes, which are the master regulators of SE. Thus, DNA methylation, seems to control SE induction and expression of the TF genes that are essential for the embryogenic transition. Identifying the SE-specific pattern of DNA methylation within these genes will contribute to deciphering the SE-associated DNA methylome.
However, it recently became evident that DNA methylation appears to control the expression of only a small number of genes (Seymour and Becker 2017) and that the impact of DNA methylation on gene expression is distinctly influenced by other epigenetic modifications including histone modification and gene silencing by small noncoding RNA (Stroud et al. 2014; Du et al. 2015; Niederhuth and Schmidt 2017). Thus, to fully understand the function of DNA methylation in the embryogenic transition the functional outcome of DNA methylation in the context of other epigenetic changes remains to be uncovered. Moreover, it is of importance to reveal the relation of DNA methylation with the recently identified miRNA that plays a regulatory role in the SE of Arabidopsis (Wójcik and Gaj 2016; Szyrajew et al. 2017; Wójcik et al. 2017).
References
Aceituno FF, Moseyko N, Rhee SY, Gutiérrez R (2008) The rules of gene expression in plants: organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana. BMC Genom 9:438. https://doi.org/10.1186/1471-2164-9-438
Ashapkin VV, Kutueva LI, Vanyushin BF (2016) Plant DNA methyltransferase genes: multiplicity, expression, methylation patterns. Biochemistry 81:141–151. https://doi.org/10.1134/S0006297916020085
Bauer MJ, Fischer RL (2011) Genome demethylation and imprinting in the endosperm. Curr Opin Plant Biol 14:162–167. https://doi.org/10.1016/j.pbi.2011.02.006
Berdasco M, Alcázar R, García-Ortiz MV, Ballestar E, Fernández AF, Roldán-Arjona T, Tiburcio AF, Altabella T, Buisine N, Quesneville H, Baudry A, Lepiniec L, Alaminos M, Rodríguez R, Lloyd A, Colot V, Bender J, Canal MJ, Esteller M, Fraga MF (2008) Promoter DNA hypermethylation and gene repression in undifferentiated Arabidopsis cells. PLoS ONE 3:e3306. https://doi.org/10.1371/journal.pone.0003306
Bewick AJ, Schmitz RJ (2017) Gene body DNA methylation in plants. Curr Opin Plant Biol 36:103–110. https://doi.org/10.1016/j.pbi.2016.12.007
Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AAM, Miki BLA, Custers JBM, van Lookeren Campagne MM (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14:1737–1749. https://doi.org/10.1105/tpc.001941
Bouyer B, Kramdi A, Kassam K, Heese M, Schnittger A, Roudier F, Colot V (2017) DNA methylation dynamics during early plant life. Genome Biol 18:179. https://doi.org/10.1186/s13059-017-1313-0
Braybrook SA, Harada JJ (2008) LECs go crazy in embryo development. Trends Plant Sci 13:624–630. https://doi.org/10.1016/j.tplants.2008.09.008
Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RL, Harada JJ (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA 103:3468–3473. https://doi.org/10.1073/pnas.0511331103
Cao X, Jacobsen SE (2002a) Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12:1138–1144. https://doi.org/10.1016/S0960-9822(02)00925-9
Cao X, Jacobsen SE (2002b) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. PNAS 99(Suppl 4):16491–16498. https://doi.org/10.1073/pnas.162371599
Chan SWL, Henderson IR, Zhang X, Shah G, Chien JSC, Jacobsen SE (2006) RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in Arabidopsis. PLoS Genet 2:e83. https://doi.org/10.1371/journal.pgen.0020083
Chang S, Pikaard CS (2005) Transcript profiling in Arabidopsis reveals complex responses to global inhibition of DNA methylation and histone deacetylation. J Biol Chem 280:796–804. https://doi.org/10.1074/jbc.M409053200
Chawla R, Nicholson SJ, Folta KM, Srivastava V (2007) Transgene-induced silencing of Arabidopsis phytochrome A gene via exonic methylation. Plant J 52:1105–1118. https://doi.org/10.1111/j.1365-313X.2007.03301.x
Christman JK (2002) 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21:5483–5495. https://doi.org/10.1038/sj.onc.1205699
Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE (2008) Shotgun bisulfite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452:215–219. https://doi.org/10.1038/nature06745
De-la-Peña C, Nic-Can GI, Galaz-Ávalos RM, Avilez-Montalvo R, Loyola-Vargas VM (2015) The role of chromatin modifications in somatic embryogenesis in plants. Front Plant Sci 6:635. https://doi.org/10.3389/fpls.2015.00635
Doyle J, Doyle JL (1987) Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem Bull 19:11–15
Du J, Johnson LM, Jacobsen SE, Patel DJ (2015) DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol 16:519–532. https://doi.org/10.1038/nrm4043
Elhiti M, Stasolla C, Wang A (2013) Molecular regulation of plant somatic embryogenesis. In Vitro Cell Dev Biol–Plant 49:631. https://doi.org/10.1007/s11627-013-9547-3
Fraga HPF, Vieira LN, Caprestano CA, Steinmacher DA, Micke GA, Spudeit DA, Pescador R, Guerra MP (2012) 5-Azacytidine combined with 2,4-D improves somatic embryogenesis of Acca sellowiana (O. Berg) Burret by means of changes in global DNA methylation levels. Plant Cell Rep 31:2165–2176. https://doi.org/10.1007/s00299-012-1327-8
Gaj MD (2001) Direct somatic embryogenesis as a rapid and efficient system for in vitro regeneration of Arabidopsis thaliana. Plant Cell Tissue Organ Cult 64:39–46. https://doi.org/10.1023/A:1010679614721
Gaj MD, Zhang S, Harada JJ, Lemaux PG (2005) LEAFY COTYLEDON genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222:977–988. https://doi.org/10.1007/s00425-005-0041-y
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirement of suspension cultures of soybean root cells. Exp Cell Res 50:151–158. https://doi.org/10.1016/0014-4827(68)90403-5
Gliwicka M, Nowak K, Balazadeh S, Mueller-Roeber B, Gaj MD (2013) Extensive modulation of the transcription factor transcriptome during somatic embryogenesis in Arabidopsis thaliana. PLoS ONE 8:e69261. https://doi.org/10.1371/journal.pone.0069261
Gong Z, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, Zhu JK (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111:803–814. https://doi.org/10.1016/S0092-8674(02)01133-9
Grafi G, Ofir R, Chalifa-Caspi V, Plaschkes I (2011) Illuminating hidden features of stem cells. In: Kallos MK (ed) Embryonic stem cells: basic biology to bioengineering, INTECH Open Access Publisher, London, pp 265–282
Guo F, Liu C, Xia H, Bi Y, Zhao C, Zhao S, Hou L, Li F, Wang X (2013) Induced expression of AtLEC1 and AtLEC2 differentially promotes somatic embryogenesis in transgenic tobacco plants. PLoS ONE 8:e71714. https://doi.org/10.1371/journal.pone.0071714
He G, Elling AA, Deng XW (2011a) The epigenome and plant development. Annu Rev Plant Biol 62:411–435. https://doi.org/10.1146/annurev-arplant-042110-103806
He XJ, Chen T, Zhu JK (2011b) Regulation and function of DNA methylation in plants and animals. Cell Res 21:442–465. https://doi.org/10.1038/cr.2011.23
Heidmann I, De Lange B, Lambalk J, Angenent GC, Boutilier K (2011) Efficient sweet pepper transformation mediated by the BABY BOOM transcription factor. Plant Cell Rep 30:1107–1115. https://doi.org/10.1007/s00299-011-1018-x
Henderson IR, Jacobsen SE (2008) Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev 22:1597–1606. https://doi.org/10.1101/gad.1667808
Horstman A, Li M, Heidmann I, Weemen M, Chen B, Muiño JM, Angenent GC, Boutilier K (2017) The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol 175:848–857. https://doi.org/10.1104/pp.17.00232
Hsieh TF, Ibarra CA, Silva P, Zemach Z, Eshed-Williams L, Fischer RL, Zilberman D (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324:1451–1454. https://doi.org/10.1126/science.1172417
Huang H, Han SS, Wang Y, Zhang XZ, Han ZH (2012) Variations in leaf morphology and DNA methylation following in vitro culture of Malus xiaojinensis. Plant Cell Tissue Organ Cult 111:153–161. https://doi.org/10.1007/s11240-012-0179-9
Issa JPJ, Kantarjian HM (2009) Targeting DNA methylation. Clin Cancer Res 15:3938–3946. https://doi.org/10.1158/1078-0432.CCR-08-2783
Jeddeloh JA, Stokes TL, Richards EJ (1999) Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet 22:94–97. https://doi.org/10.1038/8803
Jiang F, Xu X, Liu H, Zhu J (2015) DRM1 and DRM2 are involved in Arabidopsis callus formation. Plant Cell Tissue Organ Cult 123:221–228. https://doi.org/10.1007/s11240-015-0812-5
Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13:484–492. https://doi.org/10.1038/nrg3230
Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F (2012) DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr Biol 22:1825–1830. https://doi.org/10.1016/j.cub.2012.07.061
Kondo H, Ozaki H, Itoh K, Kato A, Takeno K (2006) Flowering induced by 5-azacytidine, a DNA demethylating reagent in a short-day plant, Perilla frutescens var. crispa. Physiol Plant 127:130–137. https://doi.org/10.1111/j.1399-3054.2005.00635.x
Kurczyńska EU, Gaj MD, Ujczak A, Mazur E (2007) Histological analysis of direct somatic embryogenesis in Arabidopsis thaliana (L.) Heynh. Planta 226:619–628. https://doi.org/10.1007/s00425-007-0510-6
Law J, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11:204–220. https://doi.org/10.1038/nrg2719
Ledwoń A, Gaj MD (2009) LEAFY COTYLEDON2 gene expression and auxin treatment in relation to embryogenic capacity of Arabidopsis somatic cells. Plant Cell Rep 28:1677–1688. https://doi.org/10.1007/s00299-009-0767-2
Lei M, Zhanga H, Juliana R, Tanga K, Xie S, Zhu JK (2015) Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc Natl Acad Sci USA 112:3553–3557. https://doi.org/10.1073/pnas.1502279112
Leljak-Levanić D, Bauer N, Mihaljević S, Jelaska S (2004) Changes in DNA methylation during somatic embryogenesis in Cucurbita pepo L. Plant Cell Rep 23:120–127. https://doi.org/10.1007/s00299-004-0819-6
Li W, Liu H, Cheng ZJ, Su YH, Han HN, Zhang Y, Zhang XS (2011) DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet 7:e1002243. https://doi.org/10.1371/journal.pgen.1002243
Li H, Geng M, Liu Q, Jin C, Zhang Q, Chen C, Song W, Wang C (2014) Characteristics of cytosine methylation status and methyltransferase genes in the early development stage of cauliflower (Brassica oleracea L. var. botrytis). Plant Cell Tissue Organ Cult 117:187–199. https://doi.org/10.1007/s11240-014-0432-5
Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292:2077–2080. https://doi.org/10.1126/science.1059745
Liu S, Yu Y, Ruan Y, Meyer D, Wolff M, Xu L, Wang N, Steinmetz A, Shen WH (2007) Plant SET- and RING-associated domain proteins in heterochromatinization. Plant J 52:914–926. https://doi.org/10.1111/j.1365-313X.2007.03286.x
Lotan T, Ohto M, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93:1195–1205. https://doi.org/10.1016/S0092-8674(00)81463-4
Miguel C, Marum L (2011) An epigenetic view of plant cells cultured in vitro: somaclonal variation and beyond. J Exp Bot 62:3713–3725. https://doi.org/10.1093/jxb/err155
Mozgová I, Muñoz-Viana R, Hennig L (2017) PRC2 represses hormone-induced somatic embryogenesis in vegetative tissue of Arabidopsis thaliana. PLoS Genet 13:e1006562. https://doi.org/10.1371/journal.pgen.1006562
Nic-Can GI, Loyola-Vargas VM (2016) The role of the auxins during somatic embryogenesis. In: Loyola-Vargas VM, Ochoa-Alejo N (eds) Somatic embryogenesis: fundamental aspects and applications. Springer International Publishing, Switzerland, pp 171–182
Nic-Can GI, López-Torres A, Barredo-Pool F, Wrobel K, Loyola-Vargas VM, Rojas-Herrera R, De-la-Peña C (2013) New insights into somatic embryogenesis: LEAFY COTYLEDON1, BABY BOOM1 and WUSCHEL-RELATED HOMEOBOX4 are epigenetically regulated in Coffea canephora. PLoS ONE 8:e72160. https://doi.org/10.1371/journal.pone.0072160
Niederhuth CE, Schmidt RJ (2017) Putting DNA methylation in context: from genomes to gene expression in plants. Biochim Biophys Acta 1860:149–156. https://doi.org/10.1016/j.bbagrm.2016.08.009
Noceda C, Salaj T, Pérez M, Viejo M, Cañal MJ, Salaj J, Rodríguez R (2009) DNA demethylation and decrease on free polyamines is associated with the embryogenic capacity of Pinus nigra Arn. cell culture. Trees 23:1285–1293. https://doi.org/10.1007/s00468-009-0370-8
Nowak K, Gaj MD (2016) Transcription factors in the regulation of somatic embryogenesis. In: Loyola-Vargas VM, Ochoa-Alejo N (eds) Somatic embryogenesis: fundamental aspects and applications. Springer International Publishing, Switzerland, pp 53–79
Parizot B, De Rybel B, Beeckman T (2010) VisuaLRTC: a new view on lateral root initiation by combining specific transcriptome data sets. Plant Physiol 153:34–40. https://doi.org/10.1104/pp.109.148676
Parra R, Pastor MT, Pérez-Payá E, Amo-Marco JB (2001) Effect of in vitro shoot multiplication and somatic embryogenesis on 5-methylcytosine content in DNA of Myrtus communis L. Plant Growth Regul 33:131–136. https://doi.org/10.1023/A:1017571911028
Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL (2007) DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA 104:6752–6757. https://doi.org/10.1073/pnas.0701861104
Qüesta JI, Fina JP, Casati P (2013) DDM1 and ROS1 have a role in UV-B induced- and oxidative DNA damage in A. thaliana. Front Plant Sci 4:420. https://doi.org/10.3389/fpls.2013.00420
Rathore MS, Mastan SG, Agarwal PK (2014) Evaluation of DNA methylation using methylation-sensitive amplification polymorphism in plant tissues grown in vivo and in vitro. Plant Growth Regul 75:11–19. https://doi.org/10.1007/s10725-014-9926-8
Rival A, Ilbert P, Labeyrie A, Torres E, Doulbeau S, Personne A, Dussert S, Beulé T, Durand-Gasselin T, Tregear JW, Jaligot E (2013) Variations in genomic DNA methylation during the long-term in vitro proliferation of oil palm embryogenic suspension cultures. Plant Cell Rep 32:359–368. https://doi.org/10.1007/s00299-012-1369-y
Rodríguez-Sanz H, Manzanera JA, Solís MT, Gómez-Garay A, Pintos B, Risueño MC, Testillano PS (2014) Early markers are present in both embryogenesis pathways from microspores and immature zygotic embryos in cork oak, Quercus suber L. BMC Plant Biol 14:224–242. https://doi.org/10.1186/s12870-014-0224-4
Ruiz-García L, Cervera MT, Martínez-Zapater JM (2005) DNA methylation increases throughout Arabidopsis development. Planta 222:301–306. https://doi.org/10.1007/s00425-005-1524-6
Santos D, Fevereiro P (2002) Loss of DNA methylation affects somatic embryogenesis in Medicago truncatula. Plant Cell Tissue Organ Cult 70:155–161. https://doi.org/10.1023/A:1016369921067
Schumann U, Lee J, Kazan K, Ayliffe M, Wang M-B (2017) DNA-demethylase regulated genes show methylation-independent spatiotemporal expression patterns. Front Plant Sci 8:1449. https://doi.org/10.3389/fpls.2017.01449
Seymour DK, Becker C (2017) The causes and consequences of DNA methylome variation in plants. Curr Opin Plant Biol 36:56–63. https://doi.org/10.1016/j.pbi.2017.01.005
Shemer O, Landau U, Candela H, Zemach A, Williams L (2015) Competency for shoot regeneration from Arabidopsis root explants is regulated by DNA methylation. Plant Sci 238:251–261. https://doi.org/10.1016/j.plantsci.2015.06.015
Shook MS, Richards EJ (2014) VIM proteins regulate transcription exclusively through the MET1 cytosine methylation pathway. Epigenetics 9:980–986. https://doi.org/10.4161/epi.28906
Solís MT, El-Tantawy AA, Cano V, Risueño MC, Testillano PS (2015) 5-azacytidine promotes microspore embryogenesis initiation by decreasing global DNA methylation, but prevents subsequent embryo development in rapeseed and barley. Front Plant Sci 6:472. https://doi.org/10.3389/fpls.2015.00472
Song Y, Liu L, Li G, An L, Tian L (2017) Trichostatin A and 5-Aza-2′-Deoxycytidine influence the expression of cold-induced genes in Arabidopsis. Plant Signal Behav 11:e1389828. https://doi.org/10.1080/15592324.2017.1389828
Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec J, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98:11806–11811. https://doi.org/10.1073/pnas.201413498
Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152:352–364. https://doi.org/10.1016/j.cell.2012.10.054
Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE (2014) Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 21:64–72. https://doi.org/10.1038/nsmb.2735
Szyf M (2009) Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol 49:243–263. https://doi.org/10.1146/annurev-pharmtox-061008-103102
Szyrajew K, Bielewicz D, Dolata J, Wójcik AM, Nowak K, Szczygieł-Sommer A, Szweykowska-Kulińska Z, Jarmołowski A, Gaj MD (2017) Micro RNAs are intensively regulated during induction of somatic embryogenesis in Arabidopsis. Front Plant Sci 8:18. https://doi.org/10.3389/fpls.2017.00018
Tariq M, Paszkowski J (2004) DNA and histone methylation in plants. Trends Genet 20:244–251. https://doi.org/10.1016/j.tig.2004.04.005
Teerawanichpan P, Krittanai P, Chauvatcharin N, Narangajavana J (2009) Purification and characterization of rice DNA methyltransferase. Plant Physiol Biochem 47:671 – 680. https://doi.org/10.1016/j.plaphy.2009.03.014
Teixeira FK, Colot V (2010) Repeat elements and the Arabidopsis DNA methylation landscape. Heredity 105:14–23. https://doi.org/10.1038/hdy.2010.52
Teyssier C, Maury S, Beaufour M, Grondin C, Delaunay A, Le Metté C, Ader K, Cadene M, Label P, Lelu-Walter MA (2014) In search of markers for somatic embryo maturation in hybrid larch (Larix × eurolepis): global DNA methylation and proteomic analyses. Physiol Plant 150:271–291. https://doi.org/10.1111/ppl.12081
Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75:291–295. https://doi.org/10.1016/S0168-1656(99)00163-7
Tokuji Y, Takano S, Tonomura M, Tanaka S, Igari K, Watanabe T (2011) Influence of 5′-azacitidine on promoting recovery of cell competence for shoot organogenesis in Arabidopsis. Plant Cell Tissue Organ Cult 106:289–297. https://doi.org/10.1007/s11240-011-9920-z
Turck F, Coupland G (2014) Natural variation in epigenetic gene regulation and its effects on plant developmental traits. Evolution 68:620–631. https://doi.org/10.1111/evo.12286
Victoria D, Aliki K, Venetia K, Georgios M, Zoe H (2018) Spatial and temporal expression of cytosine-5 DNA methyltransferase and DNA demethylase gene families of the Ricinus communis during seed development and drought stress. Plant Growth Regul 84:81. https://doi.org/10.1007/s10725-017-0323-y
Weijers D, Wagner D (2016) Transcriptional responses to the auxin hormone. Annu Rev Plant Biol 67:539–574. https://doi.org/10.1146/annurev-arplant-043015-112122
Wickramasuriya AM, Dunwell JM (2015) Global scale transcriptome analysis of Arabidopsis embryogenesis in vitro. BMC Genom 16:301. https://doi.org/10.1186/s12864-015-1504-6
Widman N, Feng S, Jacobsen SE, Pellegrini M (2014) Epigenetic differences between shoots and roots in Arabidopsis reveals tissue-specific regulation. Epigenetics 9:236–242. https://doi.org/10.4161/epi.26869
Williams BP, Pignatta D, Henikoff S, Gehring M (2015) Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLoS Genet 11:1–19. https://doi.org/10.1371/journal.pgen.1005142
Wójcik AM, Gaj MD (2016) miR393 contributes to the embryogenic transition induced in vitro in Arabidopsis via the modification of the tissue sensitivity to auxin treatment. Planta 244:231–243. https://doi.org/10.1007/s00425-016-2505-7
Wójcik AM, Nodine MD, Gaj MD (2017) miR160 and miR166/165 contribute to the LEC2-mediated auxin response involved in the somatic embryogenesis induction in Arabidopsis. Front Plant Sci 8:2024. https://doi.org/10.3389/fpls.2017.02024
Wójcikowska B, Gaj MD (2016) Somatic embryogenesis in Arabidopsis. In: Loyola-Vargas VM, Ochoa-Alejo N (eds) Somatic embryogenesis: fundamental aspects and applications. Springer International Publishing, Switzerland, pp 185–199
Wójcikowska B, Gaj MD (2017) Expression profiling of AUXIN RESPONSE FACTOR genes during somatic embryogenesis induction in Arabidopsis. Plant Cell Rep 36:843–858. https://doi.org/10.1007/s00299-017-2114-3
Wójcikowska B, Jaskóła K, Gąsiorek P, Meus M, Nowak K, Gaj M (2013) LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissue of Arabidopsis via YUCCA-mediated auxin biosynthesis. Planta 238:425–440. https://doi.org/10.1007/s00425-013-1892-2
Xiang D, Venglat P, Tibiche C, Yang H, Risseeuw E, Cao Y, Babic V, Cloutier M, Keller W, Wang E, Selvaraj G, Datla R (2011) Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Physiol 156:346–356. https://doi.org/10.1104/pp.110.171702
Xiao W, Custard KD, Brown RC, Lemmon BE, Harada JJ, Goldberg RB, Fischer RL (2006) DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell Online 18:805–814. https://doi.org/10.1105/tpc.105.038836
Xu M, Li X, Korban SS (2004) DNA-methylation alterations and exchanges during in vitro cellular differentiation in rose (Rosa hybrida L.). Theor Appl Genet 109:899–910. https://doi.org/10.1007/s00122-004-1717-6
Yamamoto N, Kobayashi H, Togashi T, Mori Y, Kikuchi K, Kuriyama K, Tokuji Y (2005) Formation of embryogenic cell clumps from carrot epidermal cells is suppressed by 5-azacytidine, a DNA methylation inhibitor. J Plant Physiol 162:47–54. https://doi.org/10.1016/j.jplph.2004.05.013
Yamamuro C, Miki D, Zheng Z, Ma J, Wang J, Yang Z, Dong J, Zhu J-K (2014) Overproduction of stomatal lineage cells in Arabidopsis mutants defective in active DNA demethylation. Nat Commun 5:4062. https://doi.org/10.1038/ncomms5062
Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153:193–205. https://doi.org/10.1016/j.cell.2013.02.033
Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SWL, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126:1189–1201. https://doi.org/10.1016/j.cell.2006.08.003
Zhang M, Kimatu JN, Xu K, Liu B (2010) DNA cytosine methylation in plant development. J Genet Genomics 37:1–12. https://doi.org/10.1016/S1673-8527(09)60020-5
Zilberman D (2017) An evolutionary case for functional gene body methylation in plants and animals. Genome Biol 18:87. https://doi.org/10.1186/s13059-017-1230-2
Acknowledgements
This work was supported in part by the research grant for The Young Scientists received by DG from the Rector of the University of Silesia in 2016.
Author information
Authors and Affiliations
Contributions
MDG conceived and designed the experiments, DG, JM and BW performed the experiments; DG, JM and MDG analysed the data and wrote the manuscript. DG and JM contributed equally to this work. All of the authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10725_2018_389_MOESM1_ESM.tif
Supplementary material 1. Fig. S1. Expression level of the DRM2 gene in the Col-0 explants cultured on the E5. Relative transcript level was normalised to the internal control (At4g27090) and calibrated to the expression level of the DRM1 gene. Values significantly different from DRM1 expression level are indicated with an asterisk (Student’s t test, P < 0.05). Error bars indicate the standard deviation (SD). (TIF 71 KB)
10725_2018_389_MOESM5_ESM.docx
Supplementary material 5. Table S4. Global changes in the DNA methylation level in the two-week-old plants of Col-0 (WT) and cmt3; drm1; drm2; drm1drm2 and drm1drm2cmt3 mutants. (DOCX 14 KB)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Grzybkowska, D., Morończyk, J., Wójcikowska, B. et al. Azacitidine (5-AzaC)-treatment and mutations in DNA methylase genes affect embryogenic response and expression of the genes that are involved in somatic embryogenesis in Arabidopsis. Plant Growth Regul 85, 243–256 (2018). https://doi.org/10.1007/s10725-018-0389-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-018-0389-1