Abstract
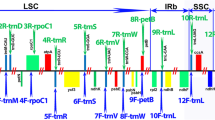
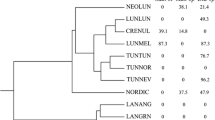
The USDA-ARS National Plant Germplasm System (NPGS) maintains a large collection of Maleae, including 49 Malus taxa, 36 Pyrus taxa, and 24 additional genera with ornamental and plant breeding value. These plant genetic resources are primarily maintained clonally as trees or shrubs in field conditions, and seeds are also conserved for some species. We used NPGS Maleae taxa to assess the genetic diversity across the tribe Maleae and placed Pyrus taxa within this broader context using analytical methods that displayed the genetic relationships as a network, rather than as a traditional dendrogram. Sequence variation from four conserved chloroplast regions unraveled the complex and often reticulate genetic relationships among and within 109 economically important Maleae taxa. In a broad sense, chloroplast haplotypes differentiated Pyrus species within Sections Pyrus and Pashia. The genetic relationships amongst Pyrus species were found to be complex, likely resulting from multiple hybridization and expansion/contraction events during the speciation process. Knowledge of the genetic relationships among Maleae genera and/or species may aid in the selection of materials for use as rootstocks and or breeding (hybridization) programs. Future collection efforts to augment the NPGS accessions within the tribe Maleae will improve the coverage and representation and assure conservation of important Rosaceae genetic resources in the NPGS.


Similar content being viewed by others
References
Aldasoro JJ, Aedo C, Muñoz Garmendia F (1996) The genus Pyrus L. (Rosaceae) in south-west Europe and North Africa. Bot J Linn Soc 121:143–158
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Campbell CS, Evans RC, Morgan DR, Dickinson TA, Arsenault MP (2007) Phylogeny of subtribe Pyrinae (formerly the Maloideae, Rosaceae): limited resolution of a complex evolutionary history. Plant Syst Evol 266:119–145
Hall T (2014) BioEdit. Biological sequence alignment editor for Win95/98/NT/2K/XP/7. http://www.mbio.ncsu.edu/bioedit/bioedit.html. Accessed 15 Sept 2014
Jiang S, Zheng X, Yu P, Yue X, Ahmed M, Cai D, Teng Y (2016) Primitive genepools of Asian pears and their complex hybrid origins inferred from fluorescent sequence-specific amplification polymorphism (SSAP) markers based on LTR retrotransposons. PLoS ONE. https://doi.org/10.1371/journal.pone.0149192
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. http://www.ub.edu/dnasp/. Accessed 15 Sept 2014
Liu J, Zheng X, Potter D, Hu C, Teng Y (2012) Genetic diversity and population structure of Pyrus calleryana (Rosaceae) in Zhejiang province, China. Biochem Syst Ecol 45:69–78
Liu J, Sun P, Zheng X, Potter D, Li K, Hu C, Teng Y (2013) Genetic structure and phylogeography of Pyrus pashia L. (Rosaceae) in Yunnan Province, China, revealed by chloroplast DNA analyses. Tree Genet Genomes 9:433–441
Liu Q, Song Y, Liu L, Zhang M, Sun J, Zhang S, Wu J (2015) Genetic diversity and population structure of pear (Pyrus spp.) collections revealed by a set of core genome-wide SSR markers. Tree Genet Genomes 11:128. https://doi.org/10.1007/211295-015-0953-z
Lo EYY, Donoghue MJ (2012) Expanded phylogenetic and dating analyses of the apples and their relatives (Pyreae, Rosaceae). Mol Phylogenet Evol 63:230–243
Medical Research Council (2014) Staden package. http://staden.sourceforge.net/. Accessed 15 Sept 2014
Nishio S, Takada N, Saito T, Yamamoto T, Iketana H (2016) Estimation of loss of genetic diversity in modern Japanese cultivars by comparison of diverse genetic resources in Asian pear (Pyrus spp.). BMC Genet 17:81. https://doi.org/10.1186/212863-016-0380-7
Paganová V (2003) Taxonomic reliability of leaf and fruit morphological characteristics of the Pyrus L. taxa in Slovakia. Hortic Sci (Prague) 30:98–107
Potter D, Gao F, Esteban Bortiri P, Oh S-H, Baggett S (2002) Phylogenetic relationships in Rosaceae inferred from chloroplast matK and trnL-trnF nucleotide sequence data. Plant Syst Evol 231:77–89
Potter D, Eriksson T, Evans RC, Oh S, Smedmark JEE, Morgan DR, Kerr M, Robertson KR, Arsenault M, Dickinson TA, Campbell CS (2007) Phylogeny and classification of Rosaceae. Plant Syst Evol 266:5–43
Raspé O, Saumitou-Laprade P, Cuguen J, Jacquemart A-L (2000) Chloroplast DNA haplotype variation and population differentiation in Sorbus aucuparia L. (Rosaceae: Maloideae). Mol Ecol 9:1113–1122
Richards CM, Volk GM, Reilley AA, Henk AD, Lockwood DR, Reeves PA, Forsline PL (2009) Genetic diversity and population structure in Malus sieversii, a wild progenitor species of domesticated apple. Tree Genet Genome 5:339–347
Robinson JP, Harris SA, Juniper BE (2001) Taxonomy of the genus Malus Mill. (Rosaceae) with emphasis on the cultivated apple, Malus domestica Borkh. Plant Syst Evol 226:35–58
Rubtsov GA (1944) Geographical distribution of the genus Pyrus and trends and factors in its evolution. Am Nat 78:358–365
Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analyses. Am J Bot 92:142–166
Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Syst Biol 49:369–381
U.S. Department of Agriculture (2017) Germplasm resources information network (GRIN-Global). GRIN taxonomy. https://npgsweb.ars-grin.gov/gringlobal/taxon/taxonomyquery.aspx. Accessed 7 Dec 2017
Velasco R et al (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42:833–841
Volk GM, Richards CM, Henk AD, Reilley AA, Bassil NV, Postman JD (2006) Diversity of wild Pyrus communis based on microsatellite analyses. J Am Soc Hortic Sci 131:408–417
Volk GM, Henk AD, Baldo A, Fazio G, Chao CT, Richards CM (2015) Chloroplast heterogeneity and historical admixture within the genus Malus. Am J Bot 102:1198–1208
Wu J, Wang Y, Xu J, Korban SS, Fei Z, Tao S et al (2018) Diversification and independent domestication of Asian and European pears. Genome Biol 19:77. https://doi.org/10.1186/s13059-018-1452-y
Xiang Y, Huang C-H, Hu Y, Wen J, Li S, Yi T, Chen H, Xiang J, Ma H (2017) Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Mol Biol Evol 34:262–281
Yu P, Jiang S, Wang X, Bai S, Teng Y (2016) Retrotransposon-based sequence-specific amplification polymorphism markers reveal that cultivated Pyrus ussuriensis originated from an interspecific hybridization. Eur J Hortic Sci 81:264–272
Zhang S-D, Jin J-J, Chen S-Y, Chase MW, Soltis DE, Li H-T, Yang J-B, Li D-Z, Yi T-S (2017) Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol 214:1355–1367
Zheng X, Cai D, Potter D, Postman J, Liu J, Teng Y (2014) Phylogeny and evolutionary histories of Pyrus L. revealed by phylogenetic trees and networks based on data from multiple DNA sequences. Mol Phylogenet Evol 80:54–65
Zong Y, Sun P, Liu J, Yue Z, Li K, Teng Y (2014a) Genetic diversity and population structure of seedling populations of Pyrus pashia. Plant Mol Biol Rep 32:644–651
Zong Y, Sun P, Liu J, Yue X, Niu Q, Teng Y (2014b) Chloroplast DNA-based genetic diversity and phylogeography of Pyrus betulaefolia (Rosaceae) in Northern China. Tree Genet Genome 10:739–749
Zong Y, Sun P, Yue X, Niu Q, Teng Y (2017) Variation in microsatellite loci reveals a natural boundary of genetic differentiation among Pyrus betulaefolia populations in Northern China. J Am Soc Hortic Sci 142:319–329
Acknowledgements
We thank Dr. Kevin Conrad at the U.S. National Arboretum for providing leaf samples for Maleae analyses and for providing an internal manuscript review. We also thank the curation teams at the Corvallis, OR and Geneva, NY repositories for providing leaf materials for genetic analyses. USDA is an equal opportunity provider and employer. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no potential conflicts of interest (financial or non-financial).
Human and animal rights
The authors have no research involving human participants and/or animals.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Volk, G.M., Henk, A.D., Richards, C.M. et al. Chloroplast sequence data differentiate Maleae, and specifically Pyrus, species in the USDA-ARS National Plant Germplasm System. Genet Resour Crop Evol 66, 5–15 (2019). https://doi.org/10.1007/s10722-018-0691-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-018-0691-9