Abstract
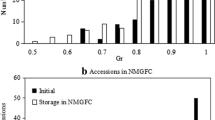
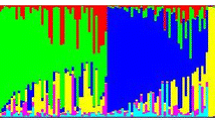
Maintaining seed viability and germplasm integrity is a challenging task in conservation of plant genetic resources, as seeds under storage will lose viability and genetic changes will occur. Attempt was made to analyze the patterns of genetic changes in wheat germplasm under ex situ genebank storage and accelerated ageing treatments. A set of 16 naturally aged wheat accessions under ex situ genebank storage since 1994 were sampled. Four recently regenerated wheat accessions were selected, four random seed samples were chosen from each accession, and three of them were exposed to three different accelerated ageing treatments. These 32 seed samples in two germplasm sets displayed a range of germination rates from 4 to 98 %. Thirty-seven microsatellite markers representing 21 wheat chromosomes were applied to screen 12 seeds of each sample and 449 SSR alleles were scored. Large SSR variation was found in each germplasm set. There was 73.1 % of the total SSR variation present among the naturally aged samples and 78.2 % present among the accelerated ageing samples. Several analyses for genetic association consistently revealed no clear genetic separations among samples of high or low germination rates in both germplasm sets. Samples under different accelerated ageing treatments did not show much genetic differentiations from the original sample of each accession. Mantel tests revealed non-significant associations between SSR variability and sample germination rates for both germplasm sets. These findings are useful for understanding seed deterioration under different ageing conditions and suggest that genome-wide SSR variability may not provide sensitive markers for the monitoring of wheat seed viability.





Similar content being viewed by others
References
Abdalla FH, Roberts EH (1968) Effects of temperature, moisture, and oxygen on the induction of chromosome damage in seeds of barley, broad beans, and peas during storage. Ann Bot 32:119–136
Arif MAR, Nagel M, Neumann K, Kobiljski B, Lohwasser U (2012) Genetic studies of seed longevity in hexaploid wheat using segregation and association mapping approaches. Euphytica 186:1–13
Association of Official Seed Analysts (1992) Seedling evaluation handbook. AOSA, Lincoln, p 98
Bednarek PT, Chwdorzewska K, Puchalski J (1998) Preliminary molecular studies on genetic changes in rye seeds due to longterm storage and regeneration. In: Gass T, Podyma W, Puchalski J, Eberhart SA (eds) Challenges in rye germplasm conservation. International Plant Genetic Resources Institute, Rome, pp 54–61
Börner A, Khlestkina EK, Chebotar S, Nagel M, Arif MAR, Neumann K, Kobiljski B, Lohwasser U, Röder MS (2012) Molecular markers in management of ex situ PGR—a case study. J Biosci 37:871–877
Boubriak I, Naumenko V, Lyne L, Osborne DJ (1999) Loss of viability in rye embryos at different levels of hydration: senescence with apoptotic nucleosome cleavage or death with random DNA fragmentation. In: Black M, Bradford KJ, Vazquez-Ramos J (eds) Seed biology: advances and applications. CABI, Cambridge, pp 205–214
Cheah KSE, Osborne DJ (1978) DNA lesions occur with loss of viability in embryos of ageing rye seeds. Nature 272:593–599
Chwedorzewska KJ, Bednarek PT, Puchalski J (2002a) AFLP profiling long term stored and regenerated rye gene bank samples. Cell Mol Biol Lett 7:457–463
Chwedorzewska KJ, Bednarek PT, Puchalski J (2002b) Studies on changes in specific rye genome regions due to seed aging and regeneration. Cell Mol Biol Lett 7:569–576
Cieslarová J, Smýkal P, Dočkalová Z, Hanáček P, Procházka S, Hýbl M, Griga M (2011) Molecular evidence of genetic diversity changes in pea (Pisum sativum L.) germplasm after long-term maintenance. Genet Resour Crop Evol 58:439–451
Dell’Aquila A (1994) Wheat seed ageing and embryo protein degradation. Seed Sci Res 4:293–298
Delouche JC, Baskin CC (1973) Accelerated aging techniques for predicting the relative storability of seed lots. Seed Sci Technol 1:427–452
Diederichsen A, Jones-Flory L (2005) Accelerated aging tests with seeds of 11 flax (Linum usitatissimum) cultivars. Seed Sci Technol 33:419–429
Donà M, Balestrazzi A, Mondoni A, Rossi G, Ventura L, Buttafava A, Macovei A, Sabatini ME, Valassi A, Carbonera D (2013) DNA profiling, telomere analysis and antioxidant properties as tools for monitoring ex situ seed longevity. Ann Bot 111:987–998
El-Maarouf-Bouteau H, Mazuy C, Corbineau F, Bailly C (2011) DNA alteration and programmed cell death during ageing of sunflower seed. J Exp Bot 62:5003–5011
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10:564–567
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
FAO (2014) Genebank standards for plant genetic resources for food and agriculture. http://www.fao.org/3/a-i3704e.pdf. Accessed 2015 November
Fu YB, Horbach C (2012) Genetic diversity in a core subset of wild barley germplasm. Diversity 4:239–257
Fu YB, Somers DJ (2009) Genome-wide reduction of genetic diversity in wheat breeding. Crop Sci 49:161–168
Fu YB, Peterson GW, Richards KW, Somers D, DePauw RM, Clarke JM (2005) Allelic reduction and genetic shift in the Canadian hard red spring wheat germplasm released from 1845 to 2004. Theor Appl Genet 110:1505–1516
Fu YB, Peterson GW, Yu J-K, Gao L, Jia L, Richards KW (2006) Impact of plant breeding on genetic diversity of the Canadian hard red spring wheat germplasm as revealed by EST derived SSR markers. Theor Appl Genet 112:1239–1247
Fu YB, Ahmed Z, Diederichsen A (2015) Towards a better monitoring of seed ageing under ex situ seed conservation. Conserv Physiol 3:cov026. doi:10.1093/conphys/cov026
Groot SPC, de Groot L, Kodde J, van Treuren R (2015) Prolonging the longevity of ex situ conserved seeds by storage under anoxia. Plant Genet Resour 13:18–26
Gutierrez G, Cruz F, Moreno J, Gonzalez-Hernandez VA, Vazquez-Ramos JM (1993) Natural and artificial seed ageing in maize: germination and DNA synthesis. Seed Sci Res 3:279–285
Hay FR, Probert RJ (2013) Advances in seed conservation of wild plant species: a review of recent research. Conserv Physiol 1:cot030. doi:10.1093/conphys/cot030
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Marcos-Filho J, McDonald MB (1998) RAPD profiles, germination and vigour of naturally and artificially aged soybean seeds. Seed Sci Technol 26:141–156
Marcos-Filho J, McDonald MB, Tekrony DM, Zhang J (1997) RAPD fragment profiles from deteriorating soybean seeds. Seed Technol 19:33–44
McDonald MB (1999) Seed deterioration: physiology, repair and assessment. Seed Sci Technol 27:177–237
Miura K, Lin SY, Yano M, Nagamine T (2002) Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.). Theor Appl Genet 104:981–986
Murata M, Roos EE, Tsuchiya T (1981) Chromosome damage induced b artificial seed ageing in barley. I. Germinability and frequency of aberrant anaphases at first mitosis. Can J Genet Cytol 23:267–280
Nagel M, Kranner I, Neumann K, Rolletschek H, Seal C, Colville L, Fernández-Marín B, Börner A (2015) Genome-wide association mapping and biochemical markers reveal that seed ageing and longevity are intricately affected by genetic background, developmental and environmental conditions in barley. Plant Cell Environ 38:1011–1022
Osborne DJ, Sharon R, Ben-Ishai R (1981) Studies on DNA integrity and DNA repair in germinating embryos of rye (Secale cereale). Isr J Bot 29:259–272
Priestley DA (1986) Seed ageing. Cornell University Press, Ithaca
Probert RJ, Daws MI, Hay FR (2009) Ecological correlates of ex situ seed longevity: a comparative study on 195 species. Ann Bot 104:57–69
Roberts EH (1973) Loss of seed viability: chromosomal and genetic aspects. Seed Sci Technol 1:515–527
Rohlf FJ (1997) NTSYS-pc 2.1. Numerical taxonomy and multivariate analysis system. Exeter Software, Setauket
Roos EE (1982) Induced genetic changes in seed germplasm during storage. In: Khan AA (ed) The physiology and biochemistry of seed development, dormancy and germination. Elsevier, New York, pp 409–434
Roos EE (1988) Genetic changes in a collection over time. Hortic Sci 23:86–90
Sackville Hamilton NR, Chorlton KH (1997) Regeneration of accessions in seed collections: a decision guide. Handbook for genebanks No. 5. International Plant Genetic Resources Institute, Rome
SAS Institute Inc. (2008) The SAS system for windows V9.2. SAS Institute Incorporated, Cary, NC, USA
Schoen DJ, David JL, Bataillon TM (1998) Deleterious mutation accumulation and the regeneration of genetic resources. Proc Natl Acad Sci USA 95:394–399
Shatters RG Jr, Schweder ME, West SH, Abdelghany A, Smith RL (1995) Environmentally induced polymorphisms detected by RAPD analysis of soybean seed DNA. Seed Sci Res 5:109–116
Sokal RR, Michener CD (1958) A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull 38:1409–1438
Swofford DL (1998) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, Massachusetts
van Treuren R, de Groot EC, van Hintum JL (2013) Preservation of seed viability during 25 years of storage under standard genebank conditions. Genet Resour Crop Evol 60:1407–1421
Vijay D, Dadlani M, Kumar PA, Panguluri SK (2009) Molecular marker analysis of differentially aged seeds of soybean and safflower. Plant Mol Biol Rep 27:282–291
Villiers TA (1973) Aging and longevity of seeds. In: Heydecker W (ed) Seed ecology. Pennsylvania State University Press, University Park, pp 265–288
Walters C, Wheeler LM, Grotenhuis JM (2005) Longevity of seeds stored in a genebank: species characteristics. Seed Sci Res 15:1–20
Zhang J, McDonald MB, Sweeney PM (1996) Random amplified polymorphic DNA (RAPD) from seeds of differing soybean and maize genotypes. Seed Sci Technol 4:513–522
Acknowledgments
Authors would like to thank Lorie Jones-Flory, Gregory Peterson, and Yasmina Bekkaoui for technical assistance in germination testing; Vicky Roslinsky for access to ABI sequencer for SSR analysis; and two anonymous journal reviewers for their helpful comments on the early version of the manuscript. This research was financially supported by an A-Base research project of Agriculture and Agri-Food Canada to YBF, and the China Scholarship Council Postdoctoral Abroad Grant to M-HY.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Fu, YB., Yang, MH., Horbach, C. et al. Patterns of SSR variation in bread wheat (Triticum aestivum L.) seeds under ex situ genebank storage and accelerated ageing. Genet Resour Crop Evol 64, 277–290 (2017). https://doi.org/10.1007/s10722-015-0349-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-015-0349-9