Abstract
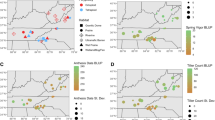
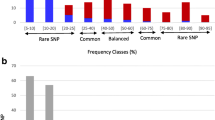
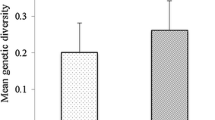
Switchgrass (Panicum virgatum), a central and Eastern USA native, is highly valued as a component in tallgrass prairie and savanna restoration and conservation projects and a potential bioenergy feedstock. The purpose of this study was to identify regional diversity, gene pools, and centers-of-diversity of switchgrass to gain an understanding of its post-glacial evolution and to identify both the geographic range and potential overlap between functional gene pools. We sampled a total of 384 genotypes from 49 accessions that included the three main taxonomic groups of switchgrass (lowland 4x, upland 4x, and upland 8x) along with one accession possessing an intermediate phenotype. We identified primary centers of diversity for switchgrass in the eastern and western Gulf Coast regions. Migration, drift, and selection have led to adaptive radiation in switchgrass, creating regional gene pools within each of the main taxa. We estimate that both upland-lowland divergence and 4x-to-8x polyploidization within switchgrass began approximately 1.5–1 M ybp and that subsequent ice age cycles have resulted in gene flow between ecotype lineages and between ploidy levels. Gene flow has resulted in “hot spots” of genetic diversity in the southeastern USA and along the Atlantic Seaboard.




Similar content being viewed by others
References
Abrams MD (1990) Adaptations and responses to drought in Quercus species of North America. Tree Physiol 7:227–238
Abrams MD (1992) Fire and the development of oak forests. Bioscience 42:246–253
Anonymous (1967) Fort Bragg history 1918–1967. http://www.mybaseguide.com/article/military/ft-bragg/615/Units-and-History
Barkworth ME, Anderton LK, Capels KM, Long S, Piep MB (2007) Manual of grasses for North America. Utah State University Press, Logan
Berger WH, Killingley JS, Vincent E (1987) Time scale of the Wisconsin/Holocene transition: oxygen isotope record in the Western Equatorial Pacific. Quatern Res 28:295–306
Bintanja R, van de Wal RSW (2008) North American ice-sheet dynamics and the onset of 100, 000-year glacial cycles. Nature 454:869–872
Buecker TR (2002) Fort Robinson and the American century, 1900–1948. Nebraska State Historical Society, Lincoln
Bunge J, Fitzpatrick M (1993) Estimating the number of species: a review. J Am Stat Assoc 88:364–373
Burnham K, Overton W (1978) Estimation of the size of a closed population when capture probabilities vary among animals. Biometrika 65:623–633
Casler MD (2005) Ecotypic variation among switchgrass populations from the northern USA. Crop Sci 45:388–398
Casler MD, Vogel KP, Taliferro CM, Wynia RL (2004) Latitudinal adaptation of switchgrass populations. Crop Sci 44:293–303
Casler MD, Stendal CA, Kapich L, Vogel KP (2007a) Genetic diversity, plant adaptation regions, and gene pools for switchgrass. Crop Sci 47:2261–2273
Casler MD, Vogel KP, Taliaferro CM, Ehlke NJ, Berdahl JD, Brummer EC, Kallenbach RL, West CP, Mitchell RB (2007b) Latitudinal and longitudinal adaptation of switchgrass populations. Crop Sci 47:2249–2260
Chao A (1987) Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43:783–791
Chao A, Hwang W, Chen Y, Kuo C (2000) Estimating the number of shared species in two communities. Stat Sin 10:227–246
Clark JS, Grimm EC, Lynch J, Mueller PG (2001) Effects of Holocene climate change on the C4 grassland/woodland boundary in the Northern Plains, USA. Ecology 82:620–636
Colwell R, Coddington J (1994) Estimating terrestrial biodiversity through extrapolation. Philos Trans R Soc Ser B 345:101–118
Cortese LM, Honig J, Miller C, Bonos SA (2010) Genetic diversity of twelve switchgrass populations using molecular and morphological markers. Bioenerg Res 3:262–271
Costich DE, Friebe B, Sheehan MJ, Casler MD, Buckler ES (2010) Genome-size variation in switchgrass (Panicum virgatum): flow cytometry and cytology reveal rampant aneuploidy. Plant Genome 3:130–141
Cwynar LC, Levesque AJ (1995) Chironomid evidence for late-glacial climatic reversals in Maine. Quatern Res 43:405–413
Deevey ES Jr (1949) Biogeography of the pleistocene: Part I: Europe and North America. Geog Soc Am Bull 60:1315–1416
Doebley J (2004) The genetics of maize evolution. Annu Rev Genet 38:37–59
Drummond A, Rambauat A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolut Biol 7:214
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Ernst WHO, Veenendaal EM, Kebakile MM (1992) Possibilities for dispersal in annual and perennial grasses in a savanna in Botswana. Vegetatio 102:1–11
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:74–75
Fay J, Fortier AC (2007) The tall grass prairie peninsula: its role in shaping American culture. Stipes Publishing, Champaign
Grimm EC, Jacobson GL Jr, Watts WA, Hansen BCS, Maasch KA (1993) A 50, 000-year record of climate oscillations from Florida and its temporal correlation with the Heinrich Events. Science 261:198–200
Gunter LE, Tuscan GA, Wullshcleger SD (1996) Diversity of switchgrass based on RAPD markers. Crop Sci 36:1017–1022
Gustafson DJ, Gibson DJ, Nickrent DL (2004) Using local seeds in prairie restoration: data support the paradigm. Native Plants Spring 2005:25–28
Harlan JR, de Wet JMJ (1975) On Ö. Winge and a prayer: the origins of polyploidy. Bot Rev 41:361–369
Hewitt G (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Bot J Linn Soc 58:247–276
Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Huang S, Siu X, Haselkorn R, Gornicki P (2003) Evolution of switchgrass (Panicum virgatum L.) based on sequences of the nuclear gene encoding plastic acetyl-CoA carboxylase. Plant Sci 164:43–49
Huang Y, Shuman B, Wang Y, Webb T III, Grimm EC, Jacobson GL Jr (2006) Climatic and environmental controls on the variation of C3 and C4 plant abundances in central Florida for the past 62, 000 years. Palaeogeog Palaeoclim Palaeoecol 237:428–435
Jacobson GT, Grimm EC (1986) A numerical analysis of Holocene forest and prairie vegetation in Central Minnesota. Ecology 67:958–966
Jakobsen BH (2009) Holocene climate change and environmental reconstruction in East Greenland. IOP Conference Series: Earth Environ Sci, vol 6, p 072028, doi:10.1088/1755-1307/6/7/072028
Kelley DW, Brachfeld SA, Nater EA, Wright HE Jr (2006) Sources of sediment in Lake Pepin on the Upper Mississippi River in response to Holocene climate changes. J Paleoclim 35:193–206
Kneller M, Peteet D (1999) Late-glacial to early Holocene climate changes from a Central Applachian pollen and macrofossil record. Quatern Res 51:133–147
Lamkey DR, Edwards JW (1999) Quantitative genetics and heterosis. In: Coors JG, Pandey S (eds) Genetics and exploration of heterosis in crops. ASA-CSSA-SSSA, Madison, pp 31–48
LaMoreaux HK, Brook GA, Knox JA (2009) Late Pleistocene and Holocene environments of the Southwestern United States from the stratigraphy and pollen content of a peat deposit on the Georgia Coastal Plain. Palaeogeog Palaeoclim Palaeoecol 280:300–312
Leigh D (2008) Late quaternary climates and river channels of the Atlantic Coastal Plain, Southeastern USA. Geomorph 101:90–108
Levesque AJ, Mayle FE, Walker IR, Cwynar LC (1993) A previously unrecognized late-glacial cold event in eastern North America. Nature 361:623–626
Maddison WP, Maddison DR (2007) Mesquite: a modular system for evolutionary analysis. Version 2.74. http://mesquiteproject.org
Martinez-Reyna JM, Vogel KP (2002) Incompatability systems in switchgrass. Crop Sci 42:1800–1805
McMillan C (1959) The role of ecotypic variation in the distribution of the central grassland of North America. Ecol Mono 29:285–308
McMillan C (1964) Ecotypic differentiation within four North American prairie grasses. I. Morphological variation within transplanted community fractions. Am J Bot 51:1119–1128
Melchinger AE (1999) Genetic diversity and heterosis. In: Coors JG, Pandey S (eds) Genetics and exploration of heterosis in crops. ASA-CSSA-SSSA, Madison, pp 99–118
Missaoui AM, Paterson AH, Bouton JH (2006) Molecular markers for the classification of switchgrass (Panicum virgatum L.) germplasm and to assess genetic diversity in three synthetic switchgrass populations. Genet Res Crop Evol 53:1291–1302
Narasimhamoorthy B, Saha MC, Swaller T, Bouton JH (2008) Genetic diversity in switchgrass collections assessed by EST-SSR markers. BioEnergy Res 1:136–146
Ocumpaugh WR, Archer S, Stuth JW (1996) Switchgrass recruitment from broadcast seed vs. seed fed to cattle. J Range Manage 49:368–371
Pakeman RJ (2001) Plant migration rates and seed dispersal mechanisms. J Biogeog 28:795–800
Palmer M (1991) Estimating species richness: the second-order jackknife reconsidered. Ecology 72:1512–1513
Peakall R, Smouse PE (2006) GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Prasad V, Stroemberg C, Alimohammadian H, Sahni A (2005) Dinosaur coprolites and the early evolution of grasses and grazers. Science 310:1170–1180
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Soltis DE, Gitzendanner MA, Strenge DD, Soltis PS (1997) Chloroplast DNA intraspecific phyolgeography of plants from the Pacific Northwest of North America. Plant Syst Evol 206:353–373
Stebbins GL (1985) Polyploidy, hybridization, and the invasion of new habitats. Ann Missouri Bot Garden 72:824–832
Stubbendieck J, Hatch SL, Butterfield CH (1991) North American range plants. University of Nebraska Press, Lincoln
Symonds VV, Soltis PS, Soltis DE (2010) Dynamics of polyploidy formation in Tragopogon (Asteraceae): recurrent formation, gene flow, and population structure. Evolution 64:1984–2003
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol (in press)
Vogel KP (2004) Switchgrass. In: Moser LE et al (eds) Warm-season (C4) grasses. ASA-CSSA-SSSA, Madison, pp 561–588
Vogel KP, Mitchell RB (2008) Heterosis in switchgrass: biomass yield in swards. Crop Sci 48:2159–2164
Vogel KP, Schmer MR, Mitchell RB (2005) Plant adaptation regions: Ecological and climatic classification of plant materials. Rangeland Ecol Manage 58:315–319
Watts WA (1971) Postglacial and interglacial vegetation history of Southern Georgia and Central Florida. Ecology 52:676–690
Watts WA, Hansen BCS, Grimm EC (1992) Camel Lake: a 40,000-yr record of vegetational and forest history from Northwest Florida. Ecology 73:1056–1066
Webb S (1986) Potential role of passenger pigeons and other vertebrates in the rapid Holocene migrations of nut trees. Quatern Res 26:367–375
Young HA, Hernlem BJ, Anderton AL, Lanzatella CL, Tobias CM (2010) Dihaploid stocks of switchgrass isolated by a screening approach. Bioenerg Res 3:305–313
Zalapa JE, Price DL, Kaeppler SM, Tobias CM, Okada M, Casler MD (2011) Hierarchical classification of switchgrass using SSR and chloroplast sequences: ecotypes, ploidies, gene pools, and cultivars. Theor Appl Genet 122:805–817
Zhong B, Yonezawa T, Zhong Y, Hasegawa M (2009) Episodic evolution and adaptation of chloroplast genomes in ancestral grasses. PLoS One 4:e5297
Acknowledgments
We thank Nick Baker, USDA-ARS, Madison, WI, and Jonathan Markham and Wesley Dean, University of Georgia, for assistance with field-plot establishment and maintenance. We thank Dr. Ken Vogel, USDA-ARS, Lincoln, NE, for many fruitful discussions, particularly his suggestion of the connection between Fort Robinson and U.S. Army bases in the eastern USA. We thank Denise Costich, USDA-ARS, Ithaca, NY, for kindly supplying a confirmed hexaploid control plant for our flow cytometry assays. We also thank Donna Tabor, Fort Bragg Historian, U.S. Army, for assistance in locating written historical records. We thank the Florida State Park Service for permission to collect switchgrass accessions on Florida State Park lands. This work was funded in part by the DOE Great Lakes Bioenergy Research Center (GLBRC, DOE Office of Science BER DE-FC02-07ER64494). Additional funding for this project was provided by the following organizations and grants: USDA-ARS CRIS Project Nos. 3655-41000-003-00D and 3655-41000-004-00D; the University of Wisconsin Agricultural Research Stations; the University of Georgia College of Agricultural and Environmental Sciences; the Ministry of Science and Technology, PR China, Project Nos. 2008BADB3B04, 2009BADA7B04, and 2011AA100209; and Project 1.3.3.3 of the DOE BioEnergy Science Center (BESC, DOE Office of Science BER DE-AC05-00OR22725). Both GLBRC and BESC are U.S. Department of Energy Bioenergy Research Centers supported by the Office of Biological and Environmental Research in the DOE Office of Science. This project represents a formal collaboration between GLBRC, BESC, and the Chinese Ministry of Science and Technology. Mention of a trademark, product name, or brand does not imply endorsement of a product over any other product by the USDA-ARS, the University of Georgia, or the U.S. Department of Energy. Panicum hallii sequence data were kindly provided to us by Eli Meyer and Tom Juenger of the University of Texas, Austin, TX. Their efforts were supported through National Science Foundation Plant Genome Research Program NSF IOS 0922457. Christian Tobias, USDA-ARS, Albany, CA kindly provided the full chloroplast sequence of P. virgatum cv. Kanlow as a reference genome for alignment of P. hallii fragments.
Author information
Authors and Affiliations
Corresponding author
Additional information
The authors Yunwei Zhang, Juan E. Zalapa and Andrew R. Jakubowski contributed equally to the work described in this manuscript.
Rights and permissions
About this article
Cite this article
Zhang, Y., Zalapa, J.E., Jakubowski, A.R. et al. Post-glacial evolution of Panicum virgatum: centers of diversity and gene pools revealed by SSR markers and cpDNA sequences. Genetica 139, 933–948 (2011). https://doi.org/10.1007/s10709-011-9597-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-011-9597-6