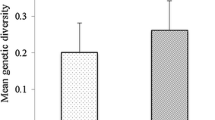
Abstract
Switchgrass (Panicum virgatum L.) is an important crop for bioenergy feedstock development. Switchgrass has two main ecotypes: the lowland ecotype being exclusively tetraploid (2n = 4x = 36) and the upland ecotype being mainly tetraploid and octaploid (2n = 8x = 72). Because there is a significant difference in ploidy, morphology, growth pattern, and zone of adaptation between and within the upland and lowland ecotypes, it is important to discriminate switchgrass plants belonging to different genetic pools. We used 55 simple sequence repeats (SSR) loci and six chloroplast sequences to identify patterns of variation between and within 18 switchgrass cultivars representing seven lowland and 11 upland cultivars from different geographic regions and of varying ploidy levels. We report consistent discrimination of switchgrass cultivars into ecotype membership and demonstrate unambiguous molecular differentiation among switchgrass ploidy levels using genetic markers. Also, SSR and chloroplast markers identified genetic pools related to the geographic origin of the 18 cultivars with respect to ecotype, ploidy, and geographical, and cultivar sources. SSR loci were highly informative for cultivar fingerprinting and to classify plants of unknown origin. This classification system is the first step toward developing switchgrass complementary gene pools that can be expected to provide a significant heterotic increase in biomass yield.



Similar content being viewed by others
References
Casler MD (2010) Changes in mean and genetic variance during two cycles of within-family selection in switchgrass. Bioenerg Res 3:47–54
Casler MD, Boe AR (2003) Cultivar × environment interactions in switchgrass. Crop Sci 43:2226–2233
Casler MD, Stendal CA, Kapich L, Vogel KP (2007a) Genetic diversity, plant adaptation regions, and gene pools for switchgrass. Crop Sci 47:2261–2273
Casler MD, Vogel KP, Taliaferro CM, Ehlke NJ, Berdahl JD, Brummer EC, Kallenbach RL, West CP, Mitchell RB (2007b) Latitudinal and longitudinal adaptation of switchgrass populations. Crop Sci 47:2249–2260
Casler MD, Mitchell RB, Vogel KP (2010) Switchgrass. In: Joshi S et al (eds) Handbook of bioenergy crops, vol 2. Taylor & Francis, New York
Cortese LM, Honig J, Miller C, Bonos SA (2010) Genetic diversity of twelve switchgrass populations using molecular and morphological markers. Bioenerg Res. doi:10.1007/s12155-010-9078-2
Costich DE, Friebe B, Sheehan MJ, Casler MD, Buckler ES (2010) Genome-size variation in switchgrass (Panicum virgatum): flow cytometry and cytology reveal rampant aneuploidy. Plant Genome (in press)
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:74–75
Gunter LE, Tuskan GA, Wullschleger SD (1996) Diversity among populations of switchgrass based on RAPD markers. Crop Sci 36:1017–1022
Hamilton MB (1999) Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Mol Ecol 8:513–525
Hopkins AA, Taliaferro CM, Murphy CD, Christian D (1996) Chromosome number and nuclear DNA content of several switchgrass populations. Crop Sci 36:1192–1195
Huang S, Su X, Haselkorn R, Gornicki P (2003) Evolution of switchgrass (Panicum virgatum L.) based on sequences of the nuclear gene encoding plastid acetyl-CoA carboxylase. Plant Sci 164:43–49
Hultquist SJ, Vogel KP, Lee DJ, Arumuganathan K, Kaeppler S (1996) Chloroplast DNA and nuclear DNA content variations among cultivars of switchgrass, Panicum virgatum L. Crop Sci 36:1049–1052
Hultquist SJ, Vogel KP, Lee DJ, Arumuganathan K, Kaeppler S (1997) DNA content and chloroplast DNA polymorphisms among switchgrasses from remnant midwestern prairies. Crop Sci 37:595–598
Lu K, Kaeppler SW, Vogel K, Arumuganathan K, Lee DJ (1998) Nuclear DNA content and chromosome numbers in switchgrass. Gt Plains Res 8:269–280
Martinez-Reyna JM, Vogel KP (2008) Heterosis in switchgrass: spaced plants. Crop Sci 48:1312–1320
Martínez-Reyna JM, Vogel KP (2002) Incompatibility systems in switchgrass. Crop Sci 42:1800–1805
McMillian C (1959) The role of ecotypic variation in the distribution of the central grassland of North America. Ecol Monogr 29:285–308
Missaoui AM, Paterson AH, Bouton JH (2006) Molecular markers for the classification of switchgrass (Panicum virgatum L.) germplasm and to assess genetic diversity in three synthetic switchgrass populations. Genet Resour Crop Evol 53:1291–1302
Narasimhamoorthy B, Saha MC, Swaller T, Bouton JH (2008) Genetic diversity in switchgrass collections assessed by EST-SSR markers. Bioenerg Res 1:136–146
Nielson EL (1944) Analysis of variation in Panicum virgatum. J Agric Res 69:327–353
Okada M, Lanzatella C, Saha MC, Bouton J, Wu R, Tobias CM (2010) Complete switchgrass genetic maps reveal subgenome collinearity, preferential pairing, and multilocus interactions. Genetics 185:745–760
Parisod C, Besnard G (2007) Glacial in situ survival in the Western Apls and polytopic autopolyploidy in Biscutella laevigata L. (Brassicaceae). Mol Ecol 16:2755–2767
Peakall R, Smouse PE (2006) GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Platt AR, Woodhall RW, George AL (2007) Improved DNA sequencing quality and efficiency using an optimized fast cycle sequencing protocol. Biotechniques 43:58–62
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Sanderson MA, Adler PR, Boateng AA, Casler MD, Sarath G (2007) Switchgrass as a biofuels feedstock in the USA. Can J Plant Sci 86:1315–1325
Shaw J, Lickey EB, Schilling EE, Small R (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare: III. Am J Bot 94:275–288
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three noncoding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Tobias C, Twigg P, Hayden D, Vogel K, Mitchell R, Lazo G, Chow EK, Sarath G (2005) Analysis of expressed sequence tags and the identification of associated short tandem repeats in switchgrass. Theor Appl Genet 111:956–964
Tobias CM, Hayden DM, Twigg P, Sarath G (2006) Genic microsatellite markers derived from EST sequences of switchgrass (Panicum virgatum L.). Mol Ecol Notes 6:185–187
Tobias CM, Sarath G, Twigg P, Lindquist E, Pangilinan J, Penning B, Barry K, Carpita N, Lazo GR (2008) Comparative genomics in switchgrass using 61,585 high-quality EST. Plant Genome 1:111–124
Vogel KP, Mitchell RB (2008) Heterosis in switchgrass: biomass yield in swards. Crop Sci 48:2159–2164
Vogel KP, Hopkins AA, Moore KJ, Johnson KD, Carlson IT (1996) Registration of ‘Shawnee’ switchgrass. Crop Sci 36:1713
Acknowledgments
The authors thank Nick Baker and Jered Giombi for their assistance with various aspects of this experiment, and Josh Hyman, Marie Adams, and Pam Prince for their invaluable help during SSR genotyping. This research was sponsored and funded by the US Department of Energy Great Lakes Bioenergy Research Center Grant DE-FC02-07ER64494.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Frisch.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zalapa, J.E., Price, D.L., Kaeppler, S.M. et al. Hierarchical classification of switchgrass genotypes using SSR and chloroplast sequences: ecotypes, ploidies, gene pools, and cultivars. Theor Appl Genet 122, 805–817 (2011). https://doi.org/10.1007/s00122-010-1488-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1488-1