Abstract
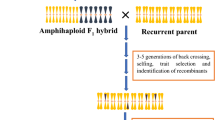
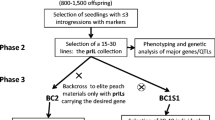
The need to boost agricultural production in the coming decades in a climate change scenario requires new approaches for the development of new crop varieties that are more resilient and more efficient in the use of resources. Crop wild relatives (CWRs) are a source of variation for many traits of interest in breeding, in particular tolerance to abiotic and biotic stresses. However, their potential in plant breeding has largely remained unexploited. CWRs can make an effective contribution to broadening the genetic base of crops and to introgressing traits of interest, but their direct use by breeders in breeding programs is usually not feasible due to the presence of undesirable traits in CWRs (linkage drag) and frequent breeding barriers with the crop. Here we call for a new approach, which we tentatively call ‘introgressiomics’, which consists of mass scale development of plant materials and populations with introgressions from CWRs into the genetic background of crops. Introgressiomics is a form of pre-emptive breeding and can be focused, when looking for specific phenotypes, or un-focused, when it is aimed at creating highly diverse introgressed populations. Exploring germplasm collections and identifying adequate species and accessions from different genepools encompassing a high diversity, using different strategies like the creation of germplasm diversity sets, Focused identification of germplasm strategy (FIGS) or gap analysis, is a first step in introgressiomics. Interspecific hybridization and backcrossing is often a major barrier for introgressiomics, but a number of techniques can be used to potentially overcome these and produce introgression populations. The generation of chromosome substitution lines (CSLs), introgression lines (ILs), or multi-parent advanced inter-cross (MAGIC) populations by means of marker-assisted selection allows not only the genetic analysis of traits present in CWRs, but also developing genetically characterized elite materials that can be easily incorporated in breeding programs. Genomic tools, in particular high-throughput molecular markers, facilitate the characterization and development of introgressiomics populations, while new plant breeding techniques (NPBTs) can enhance the introgression and use of genes from CWRs in the genetic background of crops. An efficient use of introgressiomics populations requires moving the materials into breeding pipelines. In this respect public–private partnerships (PPPs) can contribute to an increased use of introgressed materials by breeders. We hope that the introgressiomics approach will contribute to the development of a new generation of cultivars with dramatically improved yield and performance that may allow coping with the environmental changes caused by climate change while at the same time contributing to a more efficient and sustainable agriculture.



Similar content being viewed by others
References
Abberton M, Batley J, Bentley A, Bryant J, Cai H, Cockram J, Costa de Oliveira A, Cseke LJ, Dempewolf H, De Pace C, Edwards D, Gepts P, Greenland A, Hall A, Henry J, Hori K, Howe GT, Hughes S, Humphreys M, Lightfoot D, Marshall A, Mayes S, Nguyen HT, Ogbonnaya FC, Ortiz R, Paterson AH, Tuberosa R, Valliyodan B, Varshney RK, Yano M (2016) Global agricultural intensification during climate change: a role for genomics. Plant Biotechnol J 14:1095–1098. doi:10.1111/pbi.12467
Aflitos S, Schiljen E, de Jong H, de Ridder D, Smit S, Finkers R, Wang J, Zhang G, Li N, Mao L, Bakker F, Dirks R, Breit T, Gravendeel B, Huits H, Struss D, Swanson-Wagner R, van Leeuwen H, van Ham RCHJ, Fito L, Guignier L, Sevilla M, Ellul P, Ganko E, Kapur A, Reclus M, de Geus B, van de Geest H, te Lintel Hekkert B, van Haarst J, Smits L, Koops A, Sanchez-Perez G, van Heusden AW, Visser R, Quan Z, Min J, Liao L, Wang X, Wang G, Yue Z, Yang X, Xu N, Schranz E, Smets E, Vos R, Rauwerda J, Ursem R, Schuit C, Kerns M, van den Berg J, Vriezen W, Janssen A, Datema E, Jahrman T, Moquet F, Bonnet J, Peters S (2014) Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J 80:136–148. doi:10.1111/tpj.12616
Alexander LJ (1963) Transfer of a dominant type of resistance to the four known Ohio pathogenic strains of tobacco mosaic virus (TMV) from Lycopersicon peruvianum to L. esculentum. Phytopathology 53:869
Alfares W, Bouguennec A, Balfourier F, Gay G, Bergès H, Vautrin S, Sourdille P, Bernard M, Feuillet C (2009) Fine mapping and marker development for the crossability gene SKr on chromosome 5BS of hexaploid wheat (Triticum aestivum L.). Genetics 183:469–481. doi:10.1534/genetics.109.107706
Alseekh S, Ofner I, Pleban T, Tripodi P, Di Dato F, Cammareri M, Mohammad A, Grandillo S, Fernie AR, Zamir D (2013) Resolution by recombination: breaking up Solanum pennellii introgressions. Trends Plant Sci 18:536–538. doi:10.1016/j.tplants.2013.08.003
Bari A, Street K, Mackay M, Endresen DTF, De Pauw E, Amri A (2012) Focused Identification of Germplasm Strategy (FIGS) detects wheat stem rust resistance linked to environmental variables. Genet Resour Crop Evol 59:1465–1481. doi:10.1007/s10722-011-9775-5
Baute GJ, Dempewolf H, Rieseberg L (2015) Using genomic approaches to unlock the potential of CWR for crop adaptation to climate change. In: Redden R, Yadav S, Maxted N, Dulloo ME, Guarino L, Smith P (eds) Crop wild relatives and climate change. Wiley, Hoboken, pp 268–280. doi:10.1002/9781118854396.ch15
Bebber DP, Ramotowski MAT, Gurr SJ (2013) Crop pests and pathogens move polewards in a warming world. Nat Clim Change 3:985–988. doi:10.1038/nclimate1990
Bedő Z, Láng L (2015) Wheat breeding: current status and bottlenecks. In: Molnár-Láng M, Ceoloni C, Doležel J (eds) Alien introgression in wheat. Springer, Berlin Heidelberg, pp 77–101. doi:10.1007/978-3-319-23494-6_3
Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9:39. doi:10.1186/1746-4811-9-39
Bessey CE (1906) Crop improvement by utilizing wild species. J Hered os-2:112–118. doi:10.1093/jhered/os-2.1.112
Brozynska M, Furtado A, Henry RJ (2016) Genomics of crop wild relatives: expanding the gene pool for crop improvement. Plant Biotech J 14:1070–1085. doi:10.1111/pbi.12454
Campi M, Nuvolari A (2015) Intellectual property protection in plant varieties: a worldwide index (1961-2011). Res Policy 44:951–964. doi:10.1016/j.respol.2014.11.003
Cardi T (2016) Cisgenesis and genome editing: combining concepts and efforts for a smarter use of genetic resources in crop breeding. Plant Breed 135:139–147. doi:10.1111/pbr.12345
Castañeda-Alvarez NP, Khoury C, Achicanoy HA, Bernau V, Dempewolf H, Eastwood RJ, Guarino L, Harker RH, Jarvis A, Maxted N, Müller JV, Ramirez-Villegas J, Sosa CC, Struik PC, Vincent H, Toll J (2016) Global conservation priorities for crop wild relatives. Nat Plants 2:16022. doi:10.1038/nplants.2016.22
Cattivelli L, Rizza F, Badeck FW, Mazzucotelli E, Mastrangelo AM, Francia E, Marè C, Tondelli A, Stanca AM (2008) Drought tolerance improvement in crop plants: an integrated view from breeding to genomics. Field Crops Res 105:1–14. doi:10.1016/j.fcr.2007.07.004
Cavanagh C, Morell M, Mackay I, Powell W (2008) From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr Opinion Plant Biol 11:215–221. doi:10.1016/j.pbi.2008.01.002
Centro Internacional de Agricultura Tropical (2017) A global database for the distributions of crop wild relatives. doi: 10.15468/jyrthk. Accessed via http://www.gbif.org/dataset/07044577-bd82-4089-9f3a-f4a9d2170b2e on 2017-03-03
Cowling WA, Buirchell BJ, Falk DE (2009) A model for incorporating novel alleles from the primary gene pool into elite crop breeding programs while reselecting major genes for domestication or adaptation. Crop Pasture Sci 60:1009–1015. doi:10.1071/CP08223
De Storme N, Mason A (2014) Plant speciation through chromosome instability and ploidy change: cellular mechanisms, molecular factors and evolutionary relevance. Curr Plant Biol 1:10–33. doi:10.1016/j.cpb.2014.09.002
de Van Wouw M, Kik C, van Hintum T, van Treuren R, Visser B (2009) Genetic erosion in crops: concept, research results and challenges. Plant Genet Resour C 8:1–15. doi:10.1017/S1479262109990062
Dempewolf H, Hodgkins KA, Rummell SE, Ellstrand NC, Rieseberg LH (2012) Reproductive isolation during domestication. Plant Cell 24:2710–2717. doi:10.1105/tpc.112.100115
Dempewolf H, Eastwood RJ, Guarino L, Khoury C, Müller JV, Toll J (2014) Adapting agriculture to climate change: a global initiative to collect, conserve, and use crop wild relatives. Agrocecol Sust Food Syst 38:369–377. doi:10.1080/21683565.870629
Dempewolf H, Baute G, Anderson J, Kilian B, Smith C, Guarino L (2017) Past and future use of wild relatives in crop breeding. Crop Sci. doi:10.2135/cropsci2016.10.0885
Dhaliwal HS (1992) Unilateral incompatibility. In: Kalloo G, Chowdhury JB (eds) Distant hybridization of crop plants. Springer, Berlin, pp 32–46. doi:10.1007/978-3-642-84306-8_3
Díez MJ, Nuez F (2008) Tomato. In: Prohens J, Nuez F (eds) Vegetables II: Fabaceae, Liliaceae, Solanaceae, and Umbelliferae. Springer, New York, pp 249–323. doi:10.1007/978-0-387-74110-9
Dodsworth S, Chase MW, Särkinen T, Knapp S, Leitch AR (2016) Using genomic repeats for phylogenomics: a case study in wild tomatoes (Solanum section Lycopersicon: Solanaceae). Bot J Linn Soc 117:96–105. doi:10.1111/bij.12612
Dwivedi SL, Upadhyaya HD, Stalker HT, Blair MW, Bertioli DJ, Nielen S, Ortiz R (2008) Enhancing crop gene pools with beneficial traits using wild relatives. Plant Breed Rev 30:179–230. doi:10.1002/9780470380130.ch3
Fita A, Rodríguez-Burruezo A, Boscaiu M, Prohens J, Vicente O (2015) Breeding and domesticating crops adapted to drought and salinity: a new paradigm for increasing food production. Front Plant Sci 6:978. doi:10.3389/fpls.2015.00978
Friebe B, Jiang J, Raupp WJ, McIntosh RA, Gill BS (1996) Characterization of wheat-alien translocations conferring resistance to diseases and pests. Euphytica 91:59–87. doi:10.1007/BF00035277
Friebe B, Qi L, Liu C, Liu W, Gill BS (2012) Registration of a hard red winter wheat genetic stock homozygous for ph1b for facilitating alien introgression for crop improvement. J Plant Regist 6:121–123. doi:10.3198/jpr2011.05.0273crgs
Furini A, Wunder J (2004) Analysis of eggplant (Solanum melongena)-related germplasm: morphological and AFLP data contribute to phylogenetic interpretations and germplasm utilization. Theor Appl Genet 108:197–208. doi:10.1007/s00122-003-1439-1
Gerstetter C, Görlach B, Neumann K, Schaffrin D (2007) The International Treaty on Plant Genetic Resources for Food and Agriculture within the current legal regime complex on plant genetic resources. J World Intellect Prop 10:259–283. doi:10.1111/j.1747-1796.2007.00323.x
Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C (2010) Food security: the challenge of feeding 9 billion people. Science 327:812–817. doi:10.1126/science.1185383
Guerrero RF, Posto AL, Moyle LC, Hahn MW (2016) Genome-wide patterns of regulatory divergence revealed by introgression lines. Evolution 70:696–706. doi:10.1111/evo.12875
Gupta M, Mason AS, Batley J, Bharti S, Banga S, Banga SS (2016) Molecular-cytogenetic characterization of C-genome chromosome substitution lines in Brassica juncea (L.) Czern and Coss. Theor Appl Genet 129:1153–1166. doi:10.1007/s00122-016-2692-4
Gur A, Zamir D (2004) Unused natural variation can lift yield barriers in plant breeding. PLoS Biol 2:e245. doi:10.1371/journal.pbio.0020245
Gur A, Zamir D (2015) Mendelizing all components of a pyramid of three yield QTL in tomato. Front Plant Sci 6:1096. doi:10.3389/fpls.2015.01096
Haghighi KR, Ascher PD (1998) Fertile, intermediate hybrids between Phaseolus vulgaris and P. acutifolius hybrids from congruity backcrossing. Sex Plant Reprod 1:51–58. doi:10.1007/BF00227023
Hajjar R, Hodgkin T (2007) The use of crop wild relatives in crop improvement: a survey of developments over the last 20 years. Euphytica 156:1–13. doi:10.1007/s10681-007-9363-0
Hammer K (1984) Das Domestikationssyndrom. Kulturpfl 32:11–34. doi:10.1007/BF02098682
Harlan JR, de Wet JMJ (1971) Toward a rational classification of cultivated plants. Taxon 20:509–517. doi:10.2307/1218252
Hartung F, Schiemann J (2014) Precise plant breeding using new genome editing techniques: opportunities, safety and regulation in the EU. Plant J 78:742–752. doi:10.1111/tpj.12413
Herzog E, Falke KC, Presteri T, Scheuermann Ouzunova M, Frisch M (2014) Selection strategies for the development of maize introgression populations. PLoS ONE 9:e92429. doi:10.1371/journal.pone.0092429
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. Int J Clim 25:1965–1978. doi:10.1002/joc.1276
Jacobsen E, Schouten HJ (2007) Cisgenesis strongly improves introgression breeding and induced translocation breeding of plants. Trends Biotech 25:219–223. doi:10.1016/j.tibtech.2007.03.008
Jarvis DI, Hodgkin T (1999) Wild relatives and crop cultivars: detecting natural introgression and farmer selection of new genetic combinations in agroecosystems. Mol Ecol 8:S159–S173. doi:10.1046/j.1365-294X.1999.00799.x
Jarvis A, Lane A, Hijmans RI (2008) The effect of climate change on crop wild relatives. Agric Ecosyst Environ 126:13–23. doi:10.1016/j.agee.2008.01.013
Jo KR, Kim CJ, Kim SJ, Kim TY, Bergervoet M, Jongsma MA, Visser RGF, Jacobsen E, Vossen JH (2014) Development of late blight resistant potatoes by cisgene stacking. BMC Biotech 14:50. doi:10.1186/1472-6750-14-50
Johnson AAT, Veilleux RE (2000) Somatic hybridization and applications in plant breeding. Plant Breed Rev 20:167–225. doi:10.1002/9780470650189.ch6
Jones TA (2003) The restoration gene pool concept: beyond the native versus non-native debate. Restor Ecol 11:281–290. doi:10.1046/j.1526-100X.2003.00064.x
Kantar MB, Sosa CS, Khoury CK, Castañeda-Álvarez NP, Achicanoy HA, Bernau V, Kane NC, Marek L, Seiler G, Rieseberg LH (2015) Ecogeography and utility to plant breeding of the crop wild relatives of sunflower (Helianthus annuus L.). Front. Plant Sci 6:841. doi:10.3389/fpls.2015.00841
Khan MMR, Hasnunnahar M, Iwayoshi M, Ogura-Tsujira Y, Isshiki S (2015) Pollen degeneration in three functional male-sterile lines of eggplant with the wild Solanum cytoplasms. Hortic Environ Biotech 56:350–357. doi:10.1007/s13580-015-0015-3
Khazaei H, Street K, Bari A, Mackay M, Stoddard FL (2013) The FIGS (Focused Identification of Germplasm Strategy) approach identifies traits related to drought adaptation in Vicia faba genetic resources. PLoS ONE 8:e63107. doi:10.1371/journal.pone.0063107
Khlestkina EK (2014) Current applications of wheat and wheat-alien precise genetic stocks. Mol Breed 34:273–281. doi:10.1007/s11032-014-0049-8
Khoury C, Laliberté B, Guarino L (2010) Trends in ex situ conservation of plant genetic resources: a review of global crop and regional conservation strategies. Genet Resour Crop Evol 57:625–639. doi:10.1007/s10722-010-9534-z
Khush GS, Brar DS (1992) Overcoming the barriers in hybridization. In: Kalloo G, Chowdhury JB (eds) Distant hybridization of crop plants. Springer, Berlin, pp 47–61. doi:10.1007/978-3-642-84306-8_4
Kilian B, Mammen K, Millet E, Sharma R, Graner A, Salamini F, Hammer K, Özkan H (2011) Aegilops. In: Kole C (ed) Wild Crop Relatives: genomic and breeding resources, cereals. Springer, Berlin Heidelberg, pp 1–76. doi:10.1007/978-3-642-14228-4_1
King J, Grewal S, Yang C, Hubbart S, Scholefield D, Ashling S, Edwards KJ, Allen AM, Burridge A, Bloor C, Davassi A, da Silva GJ, Chalmers K, King IP (2016) A step change in the transfer of interspecific variation into wheat from Amblyopyrum muticum. Plant Biotech. doi:10.1111/pbi.12606
Knox J, Hess T, Daccache A, Wheeler T (2012) Climate change impacts on crop productivity in Africa and South Asia. Environ Res Lett 7:034032. doi:10.1088/1748-9326/7/3/034032
Kole C, Muthamilarasan M, Henry R, Edwards D, Sharma R, Abberton M, Batley J, Bentley A, Blakeney M, Bryant J, Cai H, Cakir M, Cseke LJ, Cockram J, de Oliveira AC, De Pace C, Dempewolf H, Ellison S, Gepts P, Greenland A, Hall A, Hori K, Highes S, Humphreys MW, Iorizzo M, Ismail AB, Marshall A, Mayes S, Nguyen HT, Ogbonnaya FC, Ortiz R, Paterson AH, Simon PW, Tohme J, Tuberosa R, Valliyodan B, Varshney RK, Wullschleger SD, Yano M, Prasad M (2015) Application of genomics-assisted breeding for generation of climate resilient crops: progress and prospects. Front Plant Sci 6:563. doi:10.3389/fpls.2015.00563
Lippman ZB, Semel Y, Zamir D (2007) An integrated view of quantitative trait variation using tomato interspecific introgression lines. Curr Opin Genet Dev 17:545–552. doi:10.1016/j.gde.2007.07.007
Longin CFH, Reif JC (2014) Redesigning the exploitation of wheat genetic resources. Trends Plant Sci 19:631–636. doi:10.1016/j.tplants.2014.06.012
Lusser M (2014) Workshop on public-private partnerships in plant breeding. Science and Policy Report by the Joint Research Centre of the European Commission. Publications Office of the European Union, Luxembourg. doi:10.2791/80891
Lusser M, Parisi C, Plan D, Rodríguez-Cerezo E (2011) New plant breeding techniques: state-of-the –art and prospects for commercial development. Reference Report by the Joint Research Centre of the European Commission. Publications Office of the European Union, Luxembourg. doi:10.2791/60346
Martín A, Alvarez JB, Martín LM, Barro F, Ballesteros J (1999) The development of tritordeum: a novel cereal for food processing. J Cereal Sci 30:85–95. doi:10.1006/jcrs.1998.0235
Maxted N, Kell S (2009) Establishment of a global network for the in situ conservation of crop wild relatives: Status and needs. Commission on Genetic Resources for Food and Agriculture. FAO, Rome
Maxted N, Ford-Lloyd BV, Jury S, Kell S, Scholten M (2006) Towards a definition of a crop wild relative. Biodivers Conserv 15:2673–2685. doi:10.1007/s10531-005-5409-6
Maxted N, Dulloo E, Ford-Lloyd BV, Iriondo JM, Jarvis A (2008) Gap analysis: a tool for complementary genetic conservation assessment. Divers Distrib 14:1018–1030. doi:10.1111/j.1472-4642.2008.00512.x
McIntosh RA (1992) Pre-emptive breeding to control wheat rusts. Euphytica 63:103–113. doi:10.1007/BF00023916
McKhann HI, Camilleri C, Bérard A, Bataillon T, David JL, Reboud X, Le Corre V, Caloustian C, Gut IG, Brunel D (2004) Nested core collections maximizing genetic diversity in Arabidopsis thaliana. Plant J 38:193–202. doi:10.1011/j.1365-313X.2004.02034.x
Menda N, Strickler SR, Edwards JD, Bombarely A, Dunham DM, Martin GB, Mejia L, Hutton SF, Havey MJ, Maxwell DP, Mueller LA (2014) Analysis of wild-species introgressions in tomato inbreds uncover ancestrals origins. BMC Plant Biol 14:287. doi:10.1186/s12870-014-0287-2
Meyer RS (2015) Encouraging metadata curation in the diversity seek initiative. Nat Plants 1:15099. doi:10.1038/nplants.2015.99
Meyer RS, Purugganan MD (2013) Evolution of crop species: genetics of domestication and diversification. Nature Rev Genet 14:840–852. doi:10.1038/nrg3605
Meyer RS, DuVal AE, Jensen HR (2012) Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol 196:29–48. doi:10.1111/j.1469-8137.2012.04253.x
Moore G (2015) Strategic pre-breeding for wheat improvement. Nature Plants 1:15018. doi:10.1038/nplants.2015.18
Muñoz LC, Blair MW, Duque MC, Tohme J, Roca W (2004) Introgression in common bean × tepary bean interspecific congruity-backcross lines as measured by AFLP markers. Crop Sci 44:637–645. doi:10.2135/cropsci2004.6370
Pascual L, Albert E, Sauvage C, Duangjit J, Bouchet JP, Bitton F, Desplat N, Brunel D, Le Paslier MC, Ranc N, Bruguier L, Chauchard B, Verschave P, Causse M (2016) Dissecting quantitative trait variation in the resequencing era: complementarity of bi-parental, multi-parental and association panels. Plant Sci 242:120–130. doi:10.1016/j.plantsci.2015.06.017
Peleg Z, Fahima T, Krugman T, Abbo S, Yakir D, Korol AB, Saranga Y (2009) Genomic dissection of drought resistance in durum wheat × wild emmer wheat recombinant inbreed line population. Plant Cell Environ 32:758–779. doi:10.1111/j.1365-3040.2009.01956.x
Peleman JD, van der Voort JR (2003) Breeding by design. Trends Plant Sci 8:330–334. doi:10.1016/S1360-1385(03)00134-1
Pérez-de-Castro AM, Vilanova S, Cañizares J, Pascual L, Blanca JM, Díez MJ, Prohens J, Picó B (2012) Application of genomic tools in plant breeding. Curr Genom 13:179–195. doi:10.2174/138920212800543084
Plazas M, Vilanova S, Gramazio P, Rodríguez-Burruezo A, Fita A, Herraiz FJ, Ranil R, Fonseka R, Niran L, Fonseka H, Kouassi B, Kouassi A, Kouassi A, Prohens J (2016) Interspecific hybridization between eggplant and wild relatives from different genepools. J Am Soc Hortic Sci 141:34–44
Porch TG, Beaver JS, Debouck DG, Jackson SA, Kelly JD, Dempewolf H (2013) Use of wild relatives and closely related species to adapt common bean to climate change. Agronomy 3:433–461. doi:10.3390/agronomy3020433
Prakash S, Ahuja I, Upreti HC, Kumar VD, Bhat SR, Kirti PB, Chopra VL (2001) Expression of male sterility in alloplasmic Brassica juncea with Erucastrum canariense cytoplasm and the development of a fertility restoration system. Plant Breed 120:479–482. doi:10.1046/j.1439-0523.2001.00627_x
Ramírez-Villegas J, Khoury C, Jarvis A, Debouck DG, Guarino L (2010) A gap analysis methodology for collecting crop genepools: a case study with Phaseolus beans. PLoS ONE 5:e13497. doi:10.1371/journal.pone.0013497
Ramkumar G, Madhav MS, Rama Devi SJS, Umakanth B, Pandey MK, Prasad MS, Sundaram RM, Viraktamath BC, Ravindra Babu V (2016) Identification and validation of novel alleles of rice blast resistant gene Pi54, and analysis of their nucleotide diversity in landraces and wild Oryza species. Euphytica 209:725–737. doi:10.1007/s10681-016-1666-6
Ranil RHG, Niran HML, Plazas M, Fonseka RM, Fonseka HH, Vilanova S, Andújar I, Gramazio P, Fita A, Prohens J (2015) Improving seed germination of the eggplant rootstock Solanum torvum by testing multiple factors using an orthogonal array design. Sci Hort 193:174–181. doi:10.1016/j.scienta.2015.07.030
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8:e66428. doi:10.1371/journal.pone.0066428
Rieseberg LH, Carney SE (1998) Plant hybridization. New Phytol 140:599–624. doi:10.1046/j.1469-8137.1998.00315.x
Rieseberg LH, Arias DM, Ungerer MC, Linder CR, Sinervo B (1996) The effects of mating designs on introgression between chromosomally divergent sunflower species. Theor Appl Genet 93:633–644. doi:10.1007/BF00417959
Rosenzweig C, Elliott J, Deryng D, Ruane AC, Müller C, Arneth A, Boote KJ, Folberth C, Glotter M, Khabarov N, Neumann K, Piontek F, Pugh TAM, Schmid E, Stehfest E, Yang H, Jones JW (2014) Assessing agricultural risks of climate change in the 21st century in a global gridded crop model intercomparison. Proc Natl Acad Sci USA 111:3268–3273. doi:10.1073/pnas.1222463110
Salamini F, Özkan H, Brandolini A, Schäfer-Pregl R, Martin W (2002) Genetics and geography of wild cereal domestication in the Near East. Nat Rev Genet 3:429–441. doi:10.1038/nrg817
Salinas M, Capel C, Alba JM, Mora B, Cuartero J, Fernández-Muñoz R, Lozano R, Capel J (2013) Genetic mapping of two QTL from the wild tomato Solanum pimpinellifolium L. controlling resistance against two-spotted spider mite (Tetranychus urticae Koch). Theor Appl Genet 126:83–92. doi:10.1007/s00122-012-1961-0
Savage JA, Haines DF, Holbrook NM (2015) The making of giant pumpkins: how selective breeding changed the phloem of Cucurbita maxima from source to sink. Plant Cell Environ 38:1543–1554. doi:10.1111/pce.12502
Schwarz D, Rouphael Y, Colla G, Venema JH (2010) Grafting as a tool to improve tolerance of vegetables to abiotic stresses: thermal stress, water stress and organic pollutants. Sci Hort 127:162–171. doi:10.1016/j.scienta.2010.09.016
Sharma D, Knott DR (1966) The transfer of leaf rust resistance from Agropyron to Triticum by irradiation. Can J Genet Cytol 8:137–143. doi:10.1139/g66-018
Sharma DR, Kaur R, Kumar K (1996) Embryo rescue in plants—a review. Euphytica 99:325–337. doi:10.1007/BF00022289
Shivanna KR, Bahadur B (2015) Efficacy of biotechnological approaches to raise wide sexual hybrids. In: Bahadur B, Rajam MV, Sahijram L, Krishnamurthy KV (eds) Plant Biology and biotechnology, vol II. Plant genomics and biotechnology. Springer, New Delhi, pp 347–362. doi:10.1007/978-81-322-2283-5_17
Sim SC, Van Deynze A, Stoffel K, Douches DS, Zarka D, Ganal MW, Chetelat RT, Hutton SF, Scott JW, Gardner RG, Panthee DR, Mutschler M, Myers JR, Francis DM (2012) High density SNP genotyping of tomato (Solanum lycopersicum L.) reveals patterns of genetic variation due to breeding. PLoS ONE 7:e45520. doi:10.1371/journal.pone.0045520
Smith PG (1944) Embryo culture of a tomato species hybrid. Proc Am Soc Hort Sci 44:413–416
Street K, Bari A, Mackay M, Amri A (2016) How the Focused Identification of Germplasm Strategy (FIGS) is used to mine plant genetic resources for adaptive traits. In: Maxted N, Dulloo ME, Ford-Lloyd BV (eds) Enhancing crop genepool use: capturing wild relative and landrace diversity for crop improvement. CABI, Wallingford, pp 54–65. doi:10.1079/9781780646138.0054
Syfert M, Castañeda-Álvarez NP, Khoury C, Särkinen T, Sosa CC, Achicanoy HA, Bernau V, Prohens J, Daunay MC, Knapp S (2016) Crop wild relatives of the brinjal eggplant (Solanum melongena): poorly represented in genebanks and many species at risk of extinction. Am J Bot 103:635–651. doi:10.3732/ajb.1500539
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066. doi:10.1126/science.277.5329.1063
Tanksley SD, Nelson JC (1996) Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadpated germplasm into elite breeding lines. Theor Appl Genet 92:191–203. doi:10.1007/BF00223376
Tilman D, Balzer C, Hill J, Befort BL (2011) Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci USA 108:20260–20264. doi:10.1073/pnas.1116437108
Trethowan RM, Mujeeb-Kazi A (2008) Novel germplasm resources for improving environmental stress tolerance of hexaploid wheat. Crop Sci 48:1255–1265. doi:10.2135/cropsci2007.08.0477
Valkoun JJ (2001) Wheat pre-breeding using wild progenitors. Euphytica 119:17–23. doi:10.1023/A:1017562909881
Verlaan MG, Szinay D, Hutton SF, de Jong H, Kormelink R, Visser RGF, Scott JW, Bai Y (2011) Chromosomal rearrangements between tomato and Solanum chilense hamper mapping and breeding of TYLCV resistance gene Ty-1. Plant J 68:1093–1103. doi:10.1111/j.1365-313X.2011.04762.x
Villegas D, Casadesús J, Atienza SG, Martos V, Maalouf F, Karam F, Aranjuelo I, Nogués S (2010) Tritordeum, wheat and triticale yield components under multi-local Mediterranean drought conditions. Field Crops Res 116:68–74. doi:10.1016/j.fcr.2009.11.012
Vincent H, Wiersma J, Kell S, Fielder H, Dobbie S, Castañeda-Álvarez NP, Guarino L, Eastwood R, León B, Maxted N (2013) A prioritized crop wild relative inventory to help underpin global food security. Biol Conserv 167:265–275. doi:10.1016/j.biocon.2013.08.011
Vorontsova MS, Stern S, Bohs L, Knapp S (2013) African spiny Solanum (subgenus Leptostemonum, Solanaceae): a thorny phylogenetic tangle. Bot J Linn Soc 173:176–193. doi:10.1111/boj.12053
Wall JR (1970) Experimental introgression in the genus Phaseolus. I. Effect of mating systems on interspecific gene flow. Evolution 24:356–366
Warburton ML, Crossa J, Franco J, Kazi M, Trethowan R, Rajaram S, Pfeiffer W, Zhang P, Dreisigacker S, van Ginkel M (2006) Bringing wild relatives back into the family: recovering genetic diversity in CIMMYT improved wheat germplasm. Euphytica 149:289–301. doi:10.1007/s10681-005-9077-0
Warschefsky E, Penmetsa RV, Cook DR, von Wettberg EJB (2014) Back to the wilds: tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. Am J Bot 101:1791–1800. doi:10.3732/ajb.1400116
Wendler N, Mascher M, Himmelbach A, Johnston P, Pickering R, Stein N (2015) Bulbosum to go: a toolbox to utilize Hordeum vulgare/bulbosum introgressions for breeding and beyond. Mol Plant 8:1507–1519. doi:10.1016/j.molp.2015.05.004
Wulff BBH, Moscou MJ (2014) Strategies for transferring resistance into wheat: from wide crosses to GM cassettes. Front Plant Sci 5:692. doi:10.3389/fpls.2014.00692
Zamir D (2001) Improving plant breeding with exotic genetic libraries. Nature Rev Genet 2:983–989. doi:10.1038/35103590
Zamir D, Ekstein-Michelson I, Zakay Y, Navot N, Zeidan M, Sarfatti M, Eshed Y, Harel E, Pleban T, van-Oss H, Kedar N, Rabinowitch HD, Czosnek H (1994) Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, TY-1. Theor Appl Genet 88:141–146. doi:10.1007/BF00225889
Zenkteler M (1990) In vitro fertilization and wide hybridization in higher plants. Crit Rev Plant Sci 9:267–279. doi:10.1080/07352689009382290
Acknowledgements
This work was undertaken as part of the initiative “Adapting Agriculture to Climate Change: Collecting, Protecting and Preparing Crop Wild Relatives”, which is supported by the Government of Norway. The Project is managed by the Global Crop Diversity Trust with the Millennium Seed Bank of the Royal Botanic Gardens, Kew and implemented in partnership with national and international gene banks and plant breeding institutes around the world. For further information see the project website: http://www.cwrdiversity.org/. This work has also been funded in part by European Union’s Horizon 2020 research and innovation programme under Grant agreement No 677379 (G2P-SOL) and from Spanish Ministerio de Economía y Competitividad and Fondo Europeo de Desarrollo Regional (Grant AGL2015-64755-R from MINECO/FEDER, EU). Pietro Gramazio is grateful to Universitat Politècnica de València for a pre-doctoral (Programa FPI de la UPV-Subprograma 1/2013 call) contract.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Plant Breeding: the Art of Bringing Science to Life. Highlights of the 20th EUCARPIA General Congress, Zurich, Switzerland, 29 August–1 September 2016
Edited by Roland Kölliker, Richard G. F. Visser, Achim Walter & Beat Boller
Rights and permissions
About this article
Cite this article
Prohens, J., Gramazio, P., Plazas, M. et al. Introgressiomics: a new approach for using crop wild relatives in breeding for adaptation to climate change. Euphytica 213, 158 (2017). https://doi.org/10.1007/s10681-017-1938-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-017-1938-9