Abstract
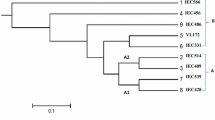
Miscanthus sinensis (M. sinensis) is a perennial C4 photosynthesis grass, with high yield, high efficiency of water usage, low fertilizer requirement, tolerance to extreme environments, and is one of the plant species with good biofuel potential. Simple sequence repeat (SSR) markers are highly informative and widely used in plant genetic studies. In this study, 88 SSR primer pairs derived from the rice genome, including 47 EST-SSRs (eSSRs) and 41 genomic SSRs (gSSRs), were evaluated for cross-species transferability to M. sinensis. Forty-one SSR primer pairs in total could successfully amplify DNA fragments in M. sinensis, showing an overall transferability rate of 46.6 % between rice and M. sinensis. The transferability rate of eSSR (51.1 %) was higher than that of gSSR (41.5 %). A total of 140 SSR loci and 340 alleles in the set of rice and M. sinensis germplasm collections were detected. Nei’s gene diversity varied from 0 to 0.72 and averaged 0.35. Shannon’s information index varied from 0 to 1.49 and averaged 0.56. The observed heterozygosity ranged from 0 to 0.95 and averaged 0.08. Thirty-nine loci (27.86 %) were shown heterozygosity out of 140 SSR loci. A dendrogram based on genetic distance showed a significant geographic differentiation. Our results indicated that 46.6% of the rice SSR markers are transferable to M. sinensis, and are useful for germplasm evaluation and genetic analysis in M. sinensis.


Similar content being viewed by others
References
Ahn S, Tanksley SD (1993) Comparative linkage maps of the rice and maize genoemes. Proc Natl Acad Sci 90(17):7980–7984
Anderson JA, Churchill GA et al (1993) Optimizing parental selection for genetic linkage maps. Genome 36(1):181–186
Bassam NE (1998) C3 and C4 plant species as energy sources and their potential impact on environment and climate. Renew Energy 15:205–210
Beale CV, Long SP (1995) Can perennial C4 grasses attain high efficiencies of radiant energy conversion in cool climates? Plant Cell Environ 18:641–650
Bennetzen JL, Freeling M (1997) The unified grass genome: synergy in synteny. Genome Res 7(4):301–306
Brenchley R, Spanning M et al (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 705(491):705–710
Chabane K, Ablett G et al (2005) EST versus genomic derived microsatellite markers for genotyping wild and cultivated barley. Genet Resour Crop Evol 52(7):903–909
Chandler VL, Wessler S (2001) Grasses: a collective model genetic system. Plant Physiol 125(3):1155–1156
Chen J, Peng J (2011) Transferability of wheat (Triticum aestivum) EST-derived SSR markers to several potential energy plants in Gramineae. Plant Sci J 29(6):696–703
Chen X, Cho YG et al (2002) Sequence divergence of rice microsatellites in Oryza and other plant species. Mol Genet Genomics 268(3):331–343
Cho YG, Ishii T et al (2000) Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.). Theor Appl Genet 100(5):713–722
Dai LJ, Wang B et al (2013) Transferability of genomic simple sequence repeat and expressed sequence tag-simple sequence repeat markers from sorghum to Miscanthus sinensis, a potential biomass crop. Crop Science. doi:10.2135/cropsci2011.12.0671
Freeling M (2001) Grasses as a single genetic system. Plant Physiol 125(3):1191–1197
Gale MD, Devos KM (1998) Plant comparative genetics after 10 years. Science 282(5389):656–659
Grass Phylogeny Working Group (2001) Phylogeny and subfamilial classfication of the Grasses (Poaceae). Ann Missouri Bot Gard 88:373–457
Greef J, Deuter M (1993) Syntaxonomy of Miscanthus x giganteus GREEF et DEU. Angewandte Botanik 67:87–90
Harushima Y, Yano M et al (1998) A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148(1):479–494
Hernández P, Dorado G et al (2001) Microsatellites and RFLP probes from maize are efficient sources of molecular markers for the biomass energy crop Miscanthus. Theor Appl Genet 102(4):616–622
Kadmon R, Pulliam HR (1993) Island biogeography: effect of geographical isolation on species composition. Ecology 74(4):978–981
Kellogg EA (1998) Relationships of cereal crops and other grasses. Proc Natl Acad Sci 95:2005–2010
Kimura M, Crow JF (1964) The number of allels that can be maintained in a finite population. Genetics 49(4):725–738
Kowalski SP, Lan TH et al (1994) Comparative mapping of Arabidopsis thaliana and Brassica oleracea chromosomes reveals islands of conserved organization. Genetics 138(2):499–510
Kurata N, Moore G et al (1994) Conservation of genome structure between rice and wheat. Nat Biotechnol 12(3):276–278
Lawrence CJ, Walbot V (2007) Translational genomics for bioenergy production from fuelstock grasses: Maize as the model species. Plant Cell 19(7):2091–2094
Lewandowski I, Clifton-Brown JC et al (2000) Miscanthus: European experience with a novel energy crop. Biomass Bioenergy 19:209–227
McCouch SR, Teytelman L et al (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9(6):199–207
Mian MAR, Saha MC et al (2005) Use of tall fescue EST-SSR markers in phylogenetic analysis of cool-season forage grasses. Genome 48(4):637–647
Morrissey JH (1981) Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem 117(2):307–310
Nei M (1972) Genetic distance between populations. Am Nat 106(949):283–292
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70(12):3321–3323
NSBC (2007) China energy statistic year book. China Statistic, Beijing
Paterson AH, Bowers JE et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457(7229):551–556
Peng JH, Lapitan NLV (2005) Characterization of EST-derived microsatellites in the wheat genome and development of eSSR markers. Funct Integr Genomics 5:80–96
Peng J, Korol AB et al (2000) Molecular genetic maps in wild emmer wheat, Triticum dicoccoides: genome-wide coverage, massive negative interference, and putative quasi-linkage. Genome Res 10(10):1509–1531
Peng J, Bai Y et al (2009) Microsatellite-based molecular diversity of bread wheat germplasm and association mapping of wheat resistance to the Russian wheat aphid. Genetica 135(1):95–122
Prince JP, Pochard E et al (1993) Construction of a molecular linkage map of pepper and a comparison of synteny with tomato. Genome 36(3):404–417
Roder MS, Plaschke J et al (1995) Abundance, variability and chromosomal location microsatellites in wheat. Mol Gen Genet 246(3):327–333
Schnable PS, Ware D et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Smith JSC, Chin ECL et al (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPS and pedigree. Theor Appl Genet 95:163–173
Somerville C, Youngs H et al (2010) Feedstocks for lignocellulosic biofuels. Science 329:790–792
Stewart CN Jr, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14(5):748–749
Tang J, Gao L et al (2006) Homologous analysis of SSR-ESTs and transferability of wheat SSR-EST markers across barley, rice and maize. Euphytica 151(1):87–93
Tang H, Bowers JE et al (2008) Synteny and collinearity in plant genomes. Science 320(5875):486–488
Varshney RK, Sigmund R et al (2005) Interspecific transferability and comparative mapping of barley EST-SSR markers in wheat, rye and rice. Plant Sci 168(1):195–202
Wang ML, Barkley NA et al (2005) Transfer of simple sequence repeat (SSR) markers from major cereal crops to minor grass species for germplasm characterization and evaluation. Plant Genet Resour 3(01):45–57
Yeh FC, Boyle TJB (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot 129:157
Yu J, Hu S et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296(5565):79–92
Zhang LY, Bernard M et al (2005) High transferability of bread wheat EST-derived SSRs to other cereals. Theor Appl Genet 111(4):677–687
Zhao H, Yu J et al (2011) Transferability of microsatellite markers from Brachypodium distachyon to Miscanthus sinensis, a potential biomass crop. J Integr Plant Biol 53:232–245
Acknowledgments
This project was supported in part by the National Natural Science Foundation of China (NSFC) Grant Nos. 30870233 and 31030055, and the Chinese Academy of Sciences under the Important Directional Program of Knowledge Innovation Project Grant No. KSCX2-YW-Z-0722.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yu, J., Zhao, H., Zhu, T. et al. Transferability of rice SSR markers to Miscanthus sinensis, a potential biofuel crop. Euphytica 191, 455–468 (2013). https://doi.org/10.1007/s10681-013-0915-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-013-0915-1