Abstract
Selection of parental lines is important in plant breeding programmes. Marker-assisted selection is an alternative to classical selection methods, which are expensive and time consuming. Marker-assisted selection aims to find molecular markers that are linked to genes that determine quantitative traits of interest. Classical statistical methods require particular assumptions to be fulfilled, which is difficult to check if the analyses are performed automatically. In this article, we present a heuristic method to find interesting markers for quantitative traits. This method includes various strategies that depend on what makes a genotype interesting to a plant breeder. This approach was applied to eighteen parental lines of winter oilseed rape F1 CMS ogura hybrids with observation of 597 markers. The traits of interest were seed yield and alkenyl glucosinolate content. Fifty-seven markers were selected for further study. The most prominent marker was OPY 02~1830. Marker-assisted selection is the first step of analysis, which can then be followed up by a more formal statistical analysis for a smaller set of interesting markers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Selection of parental lines is one of the most expensive stages of breeding hybrid varieties, but it is also important. The choice of good parent components for development of F1 hybrids strongly affects the efficiency of breeding. Until recently, the main method for selecting hybrid combinations was to evaluate field experiments based on general and specific combining ability. This method is expensive and time consuming because it requires crosses to be conducted in experimental or diallel designs. Additionally, the hybrids need to be evaluated in multiple field experiments, preferably in many environments.
The development of molecular genetics and other methods for studying genotypes at the DNA level offered an opportunity to quickly assess genetic variation in qualitative and quantitative traits independently from environmental conditions (Joudren et al. 1996; Javidfar et al. 2006). The introduction of molecular markers into breeding research revolutionised plant sciences. Molecular markers enable one to studying genotypes precisely, which was not possible with traditional methods of quantitative genetics. In addition, the results are independent of the stage of plant development or environmental conditions. Contemporary applications of molecular markers include, but are not limited to, genotyping alleles, constructing genetic maps, localising quantitative trait loci (QTLs), identifying varieties, evaluating genetic distance between hybrids or breeding lines, identifying transgenes, and monitoring gene flow (Young 1999; Mikolajczyk 2007).
Linkage groups corresponding to the chromosomes of the haploid genome were constructed with molecular markers. These linkage groups enabled researchers to determine the genetic structures of many plant and animal species. QTL mapping helped determine the localisation and function of genes that influence various traits. These techniques can be used if there is an appropriate mapping population. The integration of quantitative trait locus (QTL) analysis with marker-assisted selection (MAS) could potentially increase breeding efficiency (Steele et al. 2006; Kassahun et al. 2010). Many methods for QTL detection employ a biparental mapping population. Such investigations are highly relevant to understanding the genetic structure of plants. However, available resources limit the size of mapping populations, which results in decreased precision of QTL position and effect estimates (Dekkers and Hospital 2002; Schön et al. 2004). The main limitation of MAS is that the biparental mapping populations used in most QTL studies do not readily translate into breeding applications (Heffner et al. 2009).
Molecular markers are often helpful when a mapping population is not available. In such situations, localising genes that determine particular traits is impossible. Employment of molecular markers can help determine which markers differentiate between genotypes. This knowledge could direct decisions in breeding programmes such as choosing which parental components will be used for further crosses.
A number of methods for assessing the influence of markers on traits without mapping populations have been proposed. Early investigations with inbred lines (Soler et al. 1976) were based on statistical testing of a lack of differences between phenotypic means for groups. These groups were characterised by the presence or absence of polymorphic products of amplifications with the use of the corresponding primer. These were represented by the presence or absence of the band. A significant difference in means for the groups was interpreted as linkage between the QTL and the marker (Tanksley et al. 1982; Simpson 1989).
Linkage between molecular markers and quantitative traits without use of a mapping population was also studied through methods such as logistic regression and discriminant analysis (Mcharo et al. 2005), regression analysis (Javidfar et al. 2006; Miano et al. 2008), and two-sample comparison based on hypothesis testing (Irzykowska et al. 2005; Weber et al. 2005; Irzykowska and Bocianowski 2008). All of these methods provide parametric models based on statistical hypothesis testing, which is commonly known to strongly depend on assumptions. Violations of even one assumption can lead to inaccurate representations of the studied phenomenon. This does not constitute a problem when only a few analyses are performed because model checking techniques can be employed. However, when molecular markers are used, hundreds or thousands of analyses are necessary. In such instances, it is preferable to automate the analyses, but this is practically impossible with statistical methods that require model checking. This problem can be overcome by using biological significance instead of statistical significance (Chiplonkar and Prayag 1997; Johnson 1999; Reese 2004). Such an analysis can be performed by setting heuristic criteria that describe what one assumes to be interesting from a biological point of view; in the present context, one would derive on a set of markers that potentially influence the trait. Such an approach can also be considered as the first stage of analysis that provides interesting markers that can be further analysed with formal methods of statistical inference. Instead of analysing thousands markers through parametric methods, one can analyse a much smaller set.
Therefore, the aim of this article is to present a heuristic method for finding interesting markers in terms of a quantitative trait. The method is general and includes various strategies that depend on what makes a genotype interesting to a plant breeder. Because thousands of analyses may be required, this article also includes a discussion regarding automatically analysing associations between molecular markers and quantitative traits.
Materials and methods
Plant material and field trials
The plant material used in this study comprised eighteen parental lines of winter oilseed rape F1 CMS ogura hybrids. There were 10 paternal lines (six restorers and four without the restorer gene) and eight CMS ogura lines. Male sterile CMS ogura lines were pollinated from neighbouring plots with fertile male plants. Oilseed rape is a facultative species, and self-pollination is as good as open pollination in producing a good seed set.
Field trials were performed in four replicates of a randomised complete block design at the Experimental Station of Wielichowo in Zielęcin (52°10′N, 16°23′E) and Plant Breeding Company Strzelce Ltd in Borowo (52°07′N, 16°46′E), Poland, during the crop seasons of 2002/2003 and 2003/2004. After the harvest, the seed yield from each replicate plot (surface of the plot 10 m2) and the alkenyl glucosinolate content (gluconapin, glucobrassicanapin, progoitrin and total alkenyl glucosinolates) were measured. Analyses of glucosinolates were performed by gas chromatography of silyl derivatives of desulfoglucosinolates (Michalski et al. 1995).
The field experiment was conducted on typical loessial soil of quality class IIIa (2002, 2003) in Borowo and brown soil of quality class IVa in Zielęcin. The topsoil ranged from acidic to slightly acidic (pH 4.5–5.8). The previous crops were spring barley (2002 and 2003—Zielęcin) and winter wheat (2002 and 2003—Borowo), which are both grain crops. Winter oilseed rape seeds were sown between the 24th and 28th of August at a density of 80 plants per m2. Each plot consisted of four rows with 0.30 m between rows and approximately 0.20 m between plants within rows.
During 2002 to 2004, weather conditions were typical for Poland. However, in 2003, there were strong abiotic stresses caused by a drought from March to the end of September. The sum of rainfall in spring 2003 (100.8 mm in Borowo; 139 mm in Zielęcin) was markedly lower when compared with the same period in 2004 (176 mm in Borowo; 218 mm in Zielęcin). Average rainfall for the same period 1954–2002 was 177 mm in Borowo and 190 mm in Zielęcin.
Isozyme, RAPD, and AFLP analyses
To determine the association between different types of markers and various agronomically important characters of the parental lines of the F1 hybrids CMS ogura, analyses of RAPD, AFLP and isozyme markers were performed. Five isozyme systems were tested, including isocitrate dehydrogenase (IDH), malate dehydrogenase (MDH), 6-phosphogluconate dehydrogenase (6 PGD), leucine aminopeptidase (LAP) and phosphoglucoisomerase (PGI). All systems were tested after electrophoresis on starch gels. Extraction and electrophoretic separation of enzymatic proteins as well as staining procedures for five enzymatic systems were conducted according to methods developed by Shields et al. (1983) and Vallejos (1983).
Genomic DNA from eight-day-old leaves of 10 plants for each parental line was extracted using a modified CTAB procedure according to Doyle and Doyle (1990). The basic RAPD reaction was performed as described by Williams et al. (1990). Briefly, a total reaction volume of 12.5 μl contained 12.5 ng genomic DNA, 0.2 mM of random primer (Operon Technologies, Alameda, CA, USA), 0.1 mM dNTP mixture (MBI Fermentas Life Sciences, Burlington, ON, Canada), and 0.4 U of Taq DNA Polymerase (MBI Fermentas). An Eppendorf Mastercycler Gradient thermocycler was used to amplify the DNA fragments. The thermocycler was programmed as follows: 94°C/30 s followed by 45 cycles of 94°C/30 s, 35°C/1 min, 72°C/2 min 30 s and a final extension at 72°C for 5 min. The amplified products were separated by electrophoresis for 3 h in 1.8% agarose gels containing TBE buffer and visualised under UV light after staining with ethidium bromide. Each sample was analysed in two replicates. The DNA samples were analysed using 64 primers (Operon Technologies, USA). A majority of these primers were selected from the linkage map for rapeseed previously described by Lombard and Delourme (2001). The most important step was to identify primers generating polymorphic, distinct and clearly detectable amplification products. Only bands that were repeatedly classified as either intense or medium were included for further analysis.
AFLP analysis was performed using standard methods in accordance with the manufacturer’s instructions (Gibco BRL, AFLP Analysis Reagent Kit, AFLP Analysis System I) and as previously described by Vos et al. (1995). DNA (150 ng) was doubled-digested with the EcoRI and MseI restriction enzymes and then ligated with adaptors (Gibco BRL, AFLP Core Reagent Kit). Preamplification reactions were performed with preamplification primers carrying a single selective nucleotide at the 3′ end (+1-preamplification) (Gibco BRL AFLP Starter Primer Kit). The pre-selective PCR product was diluted at a ratio of 1:10 with TE buffer and used as a template for selective amplification. Selective AFLP amplification was performed with three selective nucleotides at the 3′ ends (EcoRI + 3 nucleotides, Mse + 3 nucleotides) (Gibco BRL AFLP Starter Primer Kit). The selective nucleotides for the AFLP primers used in this study included six EcoRI primers (E1-AAC, E2-AAG, E3-ACC, E4-ACT, E5-AGG, E6-ACA) and seven MseI primers (M1-CAA, M2-CAC, M3-CAG, M4-CAT, M5-CTA, M6-CTC, M7-CTT) in 23 primer combinations. All PCR reactions were performed in an Eppendorf Mastercycler Gradient Thermal cycler. PCR products were resolved by 13.35% denaturing polyacrylamide gels, and the gels were stained with silver. AFLP markers were named using a code for each EcoRI and MseI primer (e.g., E1M3) (Table 1), which was followed by the length of the fragment (in base pairs).
Statistical analysis
Three-way analysis of variance was conducted to determine the effects of genotype, year, location and two- and three-order interactions for the analysed traits. Analysis of variance of the data was performed using the statistical package GenStat v. 7.1. (Payne et al. 2003).
The heuristic approach proposed in this article may be based on various criteria. Examples are given below for a trait for which a better group has a bigger value. Revising the criteria for the opposite situation does not cause problems. “Two groups” refers to groups of genotypes with and without the band for the marker being considered. Examples of such criteria include the following:
-
Median (or mean) of the trait in one group exceeds that of another group by a given factor (can be given in percentages).
-
Median (or mean) of the trait in one group exceeds that of another group by some value.
-
None of the genotypes in the group exceeds a threshold value for a trait (in a biological sense).
These criteria can be used separately and in combination. Studying differences in the mean between the groups can be conducted through regular linear regression (in which case the regression slope indicates the mean difference between the groups), but testing would require that regular regression assumptions (including the homogeneity of within-group variances) be fulfilled. One can, however, use a heuristic approach in which one compares the means, thereby ignoring the possibility of violation of heterogeneous variances. Working with medians is better because they are resistant to outliers. On the other hand, outliers (especially undesirable ones) are also important. To find these, one may use a criterion for the minimum value, which is the last criterion from the list above. Any heuristic criterion based on medians or means also ignores the possibility of heterogeneous variances. However, this does not cause problems for comparisons because one can include this additional criterion. One example is where the standard deviation (or coefficient of variation) of the trait in one group does not exceed that in another group by a given factor. Another such additional criterion might be regarding outlier values (the “negative” one, which would aim to assure that no particularly bad genotypes in this group occur).
The determination of the criterion defines how one chooses an interesting marker. In median-based criteria with a standard-deviation addendum, an interesting marker is one for which the median for one group is higher for the other, as long as this group is not too variable in comparison to the other group. Various criteria may lead to choosing different markers as interesting.
For the present article, we have employed the following multi-objective criterion to determine which markers are interesting, where A and B represent values of the two groups:
-
median A/median B > 0.25, and
-
min(A) ≥ min(A,B) + 0.2 min(A,B).
This criterion was checked separately for both the A and B groups to determine whether A or B was better than the other. We also choose only those markers for which there were at least six markers for each group. The criterion for medians was used to determine if the two groups are differentiated on average, while the criterion for the minimal values ensured that no line in the chosen group performed poorly. The analyses were performed using R (R Development Core Team 2009).
After applying such a heuristic procedure, one can employ more formal statistical inferences for the chosen markers to test whether the differences in the two groups can be considered non-random.
Results
In this study, 597 differentiating markers were detected. Eighteen of these markers (3% of all markers studied) were isozymes, 225 (33.7%) were RAPD markers, and 354 (59.3%) were AFLP markers. Among the 64 RAPD starters, there were 57 differentiated DNA of the lines studied. The size of the polymorphic DNA fragments ranged from 564 to 2,100 base pairs. On average, one starter generated four polymorphic DNA fragments. The polymorphism level was 61.0%. All AFLP starters demonstrated polymorphisms among the lines studied, and polymorphic product sizes ranged between 72 and 1,352 base pairs. There were between 3 and 31 polymorphic DNA fragments for a single starter, with an overall polymorphism level of 34.5%.
Analysis of variance showed that the main effects and all two- and three-order interactions for glucobrassicanapin, progoitrin and total alkenyl glucosinolate content were statistically significant at P ≤ 0.05. Because the effect of environment was significant for all studied traits, the search for interesting markers was conducted independently in each environment. Interesting markers were those that differentiated between the measured traits in at least three out of the four environments. In addition, selected markers for gluconapin, glucobrassicanapin, progoitrin and total alkenyl glucosinolates content were those that had a smaller group median in addition to a larger median for seed yield for a particular group.
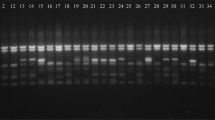
Of particular interest are the markers listed in Table 2. These markers differentiated between the studied lines in at least 8 out of 20 cases (five traits in four environments). The most prominent was marker OPY 02~1830 (Fig. 1), which showed interesting characteristics for gluconapin content in Borowo 2003, Borowo 2004 and Zielęcin 2004. It also showed interesting characteristics for glucobrassicanapin content in all four environments, total alkenyl glucosinolate content in Borowo 2004 and Zielęcin 2004, and seed yield in Borowo 2003, Borowo 2004 and Zielęcin 2004. For all 12 of these situations, lines without a band were found to be interesting. This finding means that the OPY 02~1830 marker represents a fragment of the genome that is responsible for increasing glucosinolate content, which is an undesirable anti-nutritional compound. Furthermore, marker OPY 02~1830 is characterised by relatively equal differences in comparison to other markers for median values between the groups for all studied traits (Table 2). Hence, it should be profitable to convert this dominant marker into a SCAR (sequence characterised amplified regions) marker with good reproducibility. The E2M6~1450 marker was also interesting because noticeable differences in group medians were observed. The differences in gluconapin content were 1.1, 0.4 and 0.8 in Borowo 2003, Borowo 2004 and Zielęcin 2003, respectively. For glucobrassicanapin content, the differences were 0.54, 0.42 and 0.13 in Borowo 2003, Zielęcin 2003 and Zielęcin 2004, respectively. In terms of seed yield, this marker did not differentiate between the groups as strongly, although there was still a noticeable variation; the differences were −13.1 and −5.2 in Borowo 2003 and Zielęcin 2003, respectively.
An interesting group of markers included those that influenced three traits in one environment (Table 3 shows selected results). These markers also differentiated between traits studied in other environments, but they did not differentiate between all three traits in the other environments. Overall, these markers were considered interesting in seven situations, except for marker E5M2 ~ 1650, which was only interesting in six situations. Table 3 does not include markers already listed in Table 2. Seven of the nine cases in Table 3 were from Borowo 2003. Borowo 2004 and Zielęcin 2003 had only one marker each. Five markers differentiated between gluconapin levels, glucobrassicanapin levels and seed yield. Two markers differentiated between glucobrassicanapin, progoitrin and seed yield. One marker differentiated between gluconapin, progoitrin and seed yield, and one marker differentiated between gluconapin, glucobrassicanapin and progoitrin.
There were 31 markers that differentiated the lines for glucobrassicanapin content for all four environments studied (Table 4). In 17 situations, the presence of the band represented an increase in glucobrassicanapin content, while in 11 situations, it represented a decrease. In three cases, the results were ambiguous. The presence of the band for markers E1M4 ~ 465 and E3M3 ~ 1650 represented a decrease in Borowo 2004, while in the three other environments, it represented an increase (Fig. 2). The opposite scenario was observed for marker E3M3 ~ 500. Similar trends as those reported in Table 2 were observed for the markers listed in Table 4. Greater differences in medians were found in 2003 than in 2004 for both locations. In addition, comparing the results for Borowo and Zielęcin in 2003 showed that in 20 out of 31 cases, there was a greater difference in medians between the groups in Borowo than in Zielęcin (Table 4).
Five markers (E1M5 ~ 1100, E2M2 ~ 240, OPF 20 ~ 1160, E2M7 ~ 830, and OPN 02 ~ 1500) differentiated seed yield in all four environments (Table 5). For the first three markers, the group with the band was considered positive because these genotypes had higher seed yield than those without the band. Genotypes with the band for markers E2M7 ~ 830 and OPN 02 ~ 1500 had a smaller seed yield.
Discussion
The main aim of this research was to demonstrate a heuristic method of searching for molecular markers that are linked to genes that determine quantitative traits. For analyses that need to be performed automatically, it is an alternative method to one based on statistical hypothesis testing, which requires particular assumptions to be fulfilled. If the assumptions are not fulfilled, decisions based on such a test could be wrong (Kozak 2009). In addition, sample size may have quite an impact on results of hypothesis testing (Quinn and Keough 2002; see also Kozak 2008 for a discussion concerning the correlation, but the same can be said regarding two-group comparisons). Therefore, all of the assumptions must be checked. For this problem, the most natural solution is a t-test for two-group comparisons. Although its two versions (for equal and non-equal within-group variances) assume that homogeneous within-group variances are not a problem from a statistical point of view, one still needs to assume normality. In our data, this assumption was often violated, in many cases because of skewness and/or outliers. In addition, the traits in the groups may have different variances (as mentioned, this is usually no problem from a statistical point of view), which can cause problems in interpretation if these differences are large. Additionally, there are median-based tests; however, the problem with interpreting results with large variances remains the same. Hence, every analysis requires attention. If there are thousands of analyses to carry out, such as in the case of the problem are approaching, this needs to be done automatically. In our opinion, which is also based on unpublished experiments, automatic hypothesis testing fails to detect many interesting markers. Additionally, we have described the easiest statistical approaches for the problem considered—a more advanced model would also include environmental effects. The analysis would become even more necessary and practically impossible to perform automatically. For these reasons, a heuristic approach seems to be an efficient alternative.
The aim of the proposed approach is to select “interesting” markers in terms of their associations with traits in several environments. What makes a marker interesting depends on the aims of a breeding programme, and this is determined by a plant breeder. If a marker is linked to gene which determines a trait, its band differentiates a phenotypic trait. Genotypes for which a band is observed have different values of the trait than genotypes without a band. Such an approach is general, and the only assumption it makes is that the criteria chosen can be automatically employed. This is because most interesting applications will have potentially thousands of analyses to perform. Conducting each such analysis separately would be too time-consuming.
For our analysis, we decided to compare trait medians of two groups (with or without a band) together with determining the minimum values for the groups. This approach has several advantages. The median, as a measure of central tendency, is robust to outliers, which prevents outliers from affecting the comparison as they would if mean values had been used. However, some outliers may be undesirable, so we added a criterion for the minimum value to prevent groups with undesirable outlier(s) from being selected. As mentioned in the “Materials and methods” section, one can add other criteria, such as using limits—either lower or upper, depending on the trait—for the trait’s value.
Studying the influence of markers on plant traits can provide important information for plant breeders. The method proposed is simple and can be applied at early stages of breeding programmes. In our example, we evaluated the parental forms in terms of their usefulness for further breeding. To achieve this, a mapping population is not needed even though the aim is exactly the same as that of QTL mapping: searching for markers related to plant traits. One of the important differences between these two approaches is the inability to localise the interesting marker on a linkage map. Nevertheless, such markers are sufficient for efficiently selecting genotypes with desirable traits, which is the main goal of marker-assisted selection (Martin et al. 2008; Pawłowicz et al. 2008; do Nascimento et al. 2009).
Repeatability of the results for a marker across environments suggests its usefulness in a breeding programme. For example, our analysis demonstrated approximately similar differences in medians for all environments for each trait for marker OPY 02~1830 (Table 2). For the other markers, the differences in medians between the two groups were rather different among environments, which might suggest a marker-by-environment interaction for these traits. If the markers were found in only one environment or a small subset of environments, they were not considered as interesting. Breeders are interested in finding generalisable patterns rather than those that may happen only rarely. However, an exception from this rule is when this environment or subset of environments contains interesting characteristics, such as drought or stress in general.
Markers influencing many traits can be useful in genotype selection because one such marker can be used for genotype selection for many traits. For this to occur, it is crucial that the influence of that marker on each trait has the same direction. The presence or absence of that marker band needs to be consistently positive or negative for every trait. Otherwise, the marker is characterised by an ambiguous influence on the traits. Therefore, it cannot be considered an efficient selection tool. Most markers in our study had an unambiguous influence on the traits. Their positive influence on seed yield was accompanied by their negative influence on glucosinolates content, and vice versa. However, for some markers, this type of undesirable situation was detected. For markers E1M4~465, E3M3~1650 and E3M3~500, genotypes with a band had greater trait values in some environments and smaller values in other environments when compared with genotypes without a band (Table 4). This is an undesirable result, and therefore these markers should not be considered interesting and should be removed from further breeding.
When there are only a small number of markers to analyse, there is no need to resort to automatic analyses because every marker can be analysed individually. If there are a large number of markers to analyse, our proposed heuristic procedure can be the first stage of the analysis. This method aims to find a small subset of interesting markers from among a large set of markers. The subset of interesting markers chosen can then be analysed through more formal and rigorous statistical inference.
Abbreviations
- AFLP:
-
Amplified fragment length polymorphism
- MAS:
-
Marker-assisted selection
- QTL:
-
Quantitative trait locus
- RAPD:
-
Random amplified polymorphic DNA
References
Chiplonkar SA, Prayag VR (1997) Does statistical significance always imply biological significance? Curr Sci 72:423–433
Dekkers JCM, Hospital F (2002) The use of molecular genetics in the improvement of agricultural populations. Nat Rev Genet 3:22–32
do Nascimento IR, Maluf WR, Figueira AR, Menezes CB, de Resende JTV, Faria MV, Nogueira DW (2009) Marker assisted identification of topspovirus resistant tomato genotypes in segregating progenies. Sci Agric 66(3):298–303
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Heffner EL, Sorrells ME, Jannink J-L (2009) Genomics selection for crop improvement. Crop Sci 49:1–12
Irzykowska L, Bocianowski J (2008) Genetic variation, pathogenicity and mycelial growth rate differentiation between Gaeumannomyces graminis var. tritici isolates derived from winter and spring wheat. Ann Appl Biol 152:369–375
Irzykowska L, Żółtańska E, Bocianowski J (2005) Use of molecular and conventional techniques to identify and analyze genetic variability of Rhizoctonia spp. isolates. Acta Agrobotanica 58(2):19–32
Javidfar F, Ripley VL, Roslinsky V, Zeinali H, Abdmishani C (2006) Identification of molecular markers associated with oleic and linolenic acid in spring oilseed rape (Brassica napus). Plant Breed 125:65–71
Johnson DH (1999) The insignificance of statistical significance testing. J Wildl Manag 63(3):763–772
Joudren C, Barret P, Horvais R, Delourme R, Renard M (1996) Identification of RAPD markers linked to linolenic acid genes in rapeseed. Euphytica 90:351–357
Kassahun B, Bidinger FR, Hash CT, Kuruvinashetti MS (2010) Stay-green expression in early generation sorghum [Sorghum bicolor (L.) Moench] QTL introgression lines. Euphytica 172:351–362
Kozak M (2008) Correlation coefficient and the fallacy of statistical hypothesis testing. Curr Sci 95(9):1121–1122
Kozak M (2009) Analyzing one-way experiments: a piece of cake or a pain in the neck? Sci Agricola 66(4):556–562
Lombard V, Delourme R (2001) A consensus linkage map for rapeseed (Brassica napus L.): construction and integration of three individual maps from DH populations. Theor Appl Genet 103:491–507
Martin E, Cravero V, Espósito A, López Anido F, Milanesi L, Cointry E (2008) Identification of markers linked to agronomic traits in globe artichoke. Aust J Crop Sci 1(2):43–46
Mcharo M, LaBonte DR, Clark C, Hoy M, Oard JH (2005) Molecular marker variability for southern root-knot nematode resistance in sweetpotato. Euphytica 144:125–132
Miano DW, LaBonte DR, Clark AC (2008) Identification of molecular markers associated with sweet potato resistance to sweet potato virus disease in Kenya. Euphytica 160:15–24
Michalski K, Kolodziej K, Krzymanski J (1995) Quantitative analysis of glucosinolates in seeds of oilseed rape—effect of sample preparation on analytical results. Proceedings 9th International Rapeseed Congress, vol 3. Cambridge, pp 911–913, 4–7 July 1995
Mikolajczyk K (2007) Development and practical use of DNA markers. In: Gupta S (ed) Advances in botanical research rapeseed breeding, vol 45. Academic press, London, pp 99–138
Pawłowicz I, Rapacz M, Bocianowski J (2008) Identification of AFLP markers linked with low-temperature resistance in introgressions transferred from Festuca arundinacea to Lolium multiflorum. Plant Breed Seed Sci 58:3–10
Payne R, Murrey D, Harding S, Baird D, Soutou D, Lane P (2003) GenStat for windows (7th edition)—introduction. VSN International, Oxford
Quinn GP, Keough MJ (2002) Experimental design and data analysis for biologists. Cambridge University Press, Cambridge
R Development Core Team (2009) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. http://www.R-project.org. Accessed 29 June 2010
Reese RA (2004) Does significance matter? Significance 1(1):39–40
Schön C, Utz S, Groh B, Truberg S, Openshaw S, Melchinger A (2004) QTL mapping based on resampling in a vast maize testcross experiment confirms the infinitesimal model of quantitative genetics for complex traits. Genetics 167:485–498
Shields CR, Orton CJ, Stuber CW (1983) Isozymes in plants genetics and breeding. In: Tanksley SD, Orton TJ (eds) Isozymes in plants genetics and breeding, Part A. Elsevier, Amsterdam, pp 443–458
Simpson SP (1989) Detection of linkage between quantitative trait loci and restriction fragment length polymorphisms using inbred lines. Theor Appl Genet 77:815–819
Soler M, Brody T, Genizi A (1976) On the power of experimental designs for the detection of linkage between marker loci and quantitative loci in crosses between inbred lines. Theor Appl Genet 47:35–39
Steele KA, Price AH, Shashidhar HE, Witcombe JR (2006) Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theor Appl Genet 112:208–221
Tanksley SD, Medina-Filho H, Rick CM (1982) Use of naturally-occurring enzyme variation to detect and map genes controlling quantitative traits in an interspecific backcross of tomato. Heredity 49:11–25
Vallejos CE (1983) Enzyme activity staining. In: Tanksley SD, Orton TJ (eds) Isozymes in plants genetics and breeding, Part A. Elsevier, Amsterdam, pp 469–516
Vos P, Hogers R, Sleeker M, Reijans M, Lee T, Homes M, Freiters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new concept for DNA fingerprinting. Nucl Acids Res 23:4404–4414
Weber Z, Irzykowska L, Bocianowski J (2005) Analysis of mycelial growth rates and RAPD-PCR profiles in a population of Gaeumannomyces graminis var. tritici originating from wheat plants grown from fungicide-treated seed. J Phytopathol 153:318–324
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acids Res 18:6531–6535
Young ND (1999) A cautiously optimistic vision for marker-assisted breeding. Mol Breed 5:505–510
Acknowledgment
This research was partially supported by the Ministry of Science and Higher Education, project no 3 P06A 027 25.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Bocianowski, J., Kozak, M., Liersch, A. et al. A heuristic method of searching for interesting markers in terms of quantitative traits. Euphytica 181, 89–100 (2011). https://doi.org/10.1007/s10681-011-0424-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-011-0424-z