Abstract
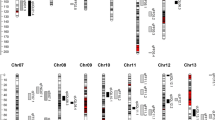
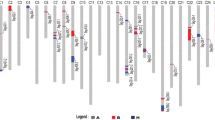
Cotton yield improvement is vital to fulfill rising global demands. The identification of major quantitative trait loci (QTL) for yield components was helpful in molecular marker-assisted selection (MAS) to improve cotton yield. We previously identified a densely populated QTL region for fiber qualities and yield components on chromosome D8 (Chro.D8) of Upland cotton from a (7235 × TM-1)RIL. In the present study, to fine-map yield component QTLs, we chose three overlapped recombinant inbred lines (RILs) with different intervals included the yield component QTLs, and backcrossed each line with TM-1 to develop three large sized mapping populations. Phenotypic data for yield components were collected in Nanjing (JES/NAU) and Xinjiang (BES/XJ) in 2006 and 2007. Three simple sequence repeat (SSR) genetic linkage maps on chro.D8 were constructed using 907 individuals in (7TR-133 × TM-1)F2 (Pop A), 670 in (7TR-132 × TM-1)F2 (Pop B), and 940 in (7TR-214 × TM-1)F2 (Pop C). Three stable QTLs for boll size, two for lint percentage and one for boll number per plant,were detected on chro.D8 following analysis of three RIL backcrossed F2/F2:3 progeny at JES/NAU and BES/XJ although their cultivation practices differ greatly between these two cotton-growing regions. One QTL for boll number per plant exhibited a phenotypic variance (PV) of 5.6–10.1%, three QTLs for boll size exhibited 15.0–35.5% PV and two lint percentage QTLs exhibited 10.9–19.3% PV. Negative correlation between lint yield and fiber strength was confirmed.

Similar content being viewed by others
References
Chen H, Qian N, Guo WZ, Song QP, Li BC, Deng FJ, Dong CG, Zhang TZ (2009) Using three overlapped RILs to dissect genetically clustered QTL for fiber strength on chro.24 in Upland cotton. Theor Appl Genet 119:605–612
Guo WZ, Cai CP, Wang CP, Han ZG, Song XL, Kai Wang, Niu XW, Wang C, Lu KY, Ben S, Zhang TZ (2007) A microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in Gossypium. Genetics 176:527–541
Jiang CX, Wright RJ, El-Zik KM, Paterson AH (1998) Polyploid formation created unique convenues for response to selection in Gossypium (cotton). Proc Acad Natl Sci USA 95:4419–4424
Kohel RJ, Richmond TR, Lewis CF (1970) Texas marker-1. Description of a genetic standard for Gossypium hirsutum L. Crop Sci 10:670–671
Lacape JM, Nguyen TB, Thibivilliers S, Bojinnov TB, Courtois B, Cantrell RG, Burr B, Hau B (2003) A combined RFLP-SSR-AFLP map of tetraploid cotton based on a Gossypium hirsutum × Gossypium barbadense backcross population. Genome 46:612–626
Lander ES, Botstein D (1986) Strategies for studying heterogeneous genetic traits in humans by using a linkage map of restriction fragment length polymorphisms. Proc Natl Acad Sci USA 83:7353–7357
McCouch SR, Cho YG, Yano PE, Blinstrub M, Morishima H, Kinoshita T (1997) Report on QTL nomenclature. Rice Genet Newslett 14:11–13
Mei M, Syed NH, Gao W, Thaxton PM, Smith CW, Stelly DM, Chen ZJ (2004) Genetic mapping and QTL analysis of fiber-related traits in cotton (Gossypium). Theor Appl Genet 108:280–291
Meredith WR Jr (1984) Quantitative genetics. In: Kohel RJ, Lewis CF (eds) Cotton. Am Soc Agro, Inc., Crop Sci Soc AM, Inc., and Soil Sci Soc AM, Inc., Madison, WI, pp 132–150
Paterson AH, Lander SE, Hewitt JD, Peterson S, Lincoln SE, Tanksely SD (1988) Resolution of quantitative traits into Mendelian factors using a complete linkage map of restriction fragement length polymorphisms. Nature 355:721–726
SAS institute (1989) SAS/STAT user’s guide version 6, 4th edn. SAS Institute, Cary, NC
Shappley ZW, Jenkins JN, Zhu J, McCarty JC (1998) Quantitative trait loci associated with agronomic and fiber traits of upland cotton. J Cotton Sci 2:153–163
Shen XL, Guo WZ, Zhu XF, Yuan YL, Yu JZ, Kohel RJ, Zhang TZ (2005) Molecular mapping of QTL for fiber qualities in three diverse lines in Upland cotton using SSR markers. Mol Breed 15:169–181
Shen XL, Guo WZ, Lu QX, Zhu XF, Yuan YL, Zhang TZ (2007) Genetic mapping of quantitative trait loci for fiber quality and yield trait by RIL approach in Upland cotton. Euphytica 155:371–380
Ulloa M, Meredith WR (2000) Genetic linkage map and QTL analysis of agronomic and fiber quality traits in an intraspecific population. J Cotton Sci 4:161–170
Ulloa M, Cantrell RG, Pency RG (2000) QTL analysis of stomatal conductance and relationship to lint yield in an interspecific cotton. J Cotton Sci 4:10–18
Ulloa M, Meredith WR, Shapplet ZW, Kahler AL (2002) RPLF genetic linkage maps from four F2:3 population and a joinmap of Gossypium hirsutum L. Thero Appl Genet 104:200–208
Ulloa M, Saha S, Jenkin N, Meredith WR, McCarty JC, Stelly DM (2005) Chromosomal assignment of RFLP linkage groups harboring important QTL on an intraspecific cotton (Gossypium hirsutum L.) joinmap. J Hered 96:132–144
Van Ooijen JW (2004) MapQTL version 5.0: software for the mapping of quantitative trait loci in experiment population. Kyazma BV, Wageningen, the Netherlands
Van Ooijen JW, Voorrips RE (2001) JoinMap version 3.0: software for the calculation of genetic linkage maps. Plant Research International, Wageningen, the Netherlands
Varshney RK, Tuberosa R (2007) Genomics-assisted crop improvement, vol. 1: Genomics approaches and platforms. Springer, Netherlands, pp 1–12
Wang K, Song XL, Han ZG, Guo WZ, Yu JZ, Sun J, Pan JJ, Kohel RJ, Zhang TZ (2006) Complete assignment of the chromosomes of G. hirsutum L. by BAC-FISH and Translocations. Theor Appl Genet 113:73–80
Acknowledgements
This work was financially supported in part by grants from the High-tech program 863 (2006AA100105), Jiangsu province key project (BE2008310, BG2006307), and the 111 Project (B08025).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, H., Qian, N., Guo, W. et al. Using three selected overlapping RILs to fine-map the yield component QTL on Chro.D8 in Upland cotton. Euphytica 176, 321–329 (2010). https://doi.org/10.1007/s10681-010-0204-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-010-0204-1