Abstract
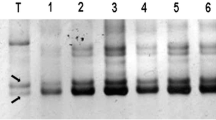
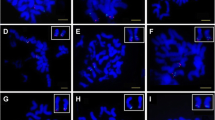
Bacterial artificial chromosome (BAC) libraries with large DNA fragment inserts have rapidly become the preferred choice for physical mapping. BAC-derived microsatellite or simple sequence repeats (SSRs) markers facilitate the integration of physical maps with genetic maps. The objective of this research was to identify chromosome locations of the BAC-derived SSR markers in tetraploid cotton. A total of 192 SSR primer pairs were derived from BAC clones of an Upland cotton genetic standard line TM-1 (Gossypium hirsutum L.). Metaphor agarose gel electrophoresis results revealed 76 and 59 polymorphic markers between TM-1 and 3–79 (G. barbadense) or G. tomentosum, respectively. Using deletion analysis method, we assigned 39 markers out of the 192 primer pairs to 17 different chromosomes or chromosome arms. Among them, 19 and 17 markers were localized to A-subgenomes (chromosome 1–13) and D-subgenomes (chromosome 14–26), respectively. The subgenome status for the remaining three markers remained unclear due to their two potential chromosome locations achieved by tertiary monosomic stocks deletion analysis. Chromosomal assignment of these BAC-derived SSR markers will help in integrating physical and cotton genetic linkage maps and thus facilitate positional candidate gene cloning, comparative genome analysis, and the coordination of chromosome-based genome sequencing project in cotton.

Similar content being viewed by others
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- EST:
-
Expressed sequence tag
- FISH:
-
Fluorescence in situ hybridization
- QTL:
-
Quantitative trait locus
- SCAR:
-
Sequence characterized amplified region
- SSR:
-
Simple sequence repeat
- SNP:
-
Single nucleotide polymorphisms
- TMB:
-
TM-1 BAC/BIBAC
- NTN:
-
Tertiary monosomic cytogenetic stock
References
Abdurakhmonov IY, Kushanov K, Djaniqulov1 F, Buriev Z, Pepper AE, Fayzieva N, Mavlonov G, Saha S, Jenkins JN, Abdukarimov A (2007) The role of induced mutation in conversion of photoperiod dependence in cotton. J Heredity 98(3):258–266
An C, Saha S, Jenkins JN, Scheffler BE, Wilkins TA, Stelly DM (2007) Transcriptome profiling, sequence characterization, and SNP-based chromosomal assignment of the EXPANSIN genes in cotton. Mol Gen Genomics 278:539–553
Blenda A, Scheffler J, Scheffler B, Palmer M, Lacape JM, Yu JZ, Jesudurai C, Jung S, Muthukumar S, Yellambalase P, Ficklin S, Staton M, Eshelman R, Ulloa M, Saha S, Burr B, Liu S, Zhang T, Fang D, Pepper A, Kumpatla S, Jacobs J, Tomkins J, Cantrell R, Main D (2006) CMD: a cotton microsatellite database resource for Gossypium genomics. BMC Genomics 7:132
Chen M, Presting G, Barbazuk WG, Goicoechea JL, Blackmon B, Fang G, Kim H, Frisch D, Yu Y, Sun S (2002) An integrated physical and genetic map of the rice genome. Plant Cell 14:537–545
Cregan PB, Mudge J, Fickus EW, Marek LF, Danish D, Denny R, Shoemaker RC, Matthews BF, Jarvik T, Young ND (1999) Targeted isolation of simple sequence repeat markers through the use of bacterial artificial chromosomes. Theor Appl Genet 98:919–928
Danesh D, Penuela S, Mudge J, Denny RL, Nordstrom H, Martinez JP, Young ND (1998) A bacterial artificial chromosome library for soybean and identification of clones near a major cyst nematode resistant gene. Theor Appl Genet 96:196–202
Frelichowski JE Jr, Palmer MB, Main D, Tomkins JP, Cantrell RG, Stelly DM, Yu J, Kohel RJ, Ulloa M (2006) Cotton genome mapping with new microsatellites from Acala ’Maxxa’ BAC-ends. Mol Gen Genomics 275:479–491
Guo W, Cai C, Wang C, Han Z, Song X, Wang K, Niu X, Wang C, Lu K, Shi B, Zhang T (2007) A microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in Gossypium. Genetics 176:527–541
Guo W, Zhang T, Shen X, Yu JZ, Kohell RJ (2003) Development of SCAR marker linked to a major QTL for high fiber strength and its usage in molecular-marker assisted selection in Upland cotton. Crop Sci 43:2252–2256
Han Z, Wang C, Song X, Guo W, Gou J, Li C, Chen X, Zhang T (2006) Characteristics, development and mapping of Gossypium hirsutum derived EST-SSRs in allotetraploid cotton. Theor Appl Genet 112:430–439
Han ZG, Guo WZ, Song XL, Zhang TZ (2004) Genetic mapping of EST-derived microsatellites from the diploid Gossypium aboreum in allotetraploid cotton. Mol Gen Genomics 272:308–327
Hanson RE, Zwick MS, Choi S, Islam-Faridi MN, McKnight TD, Wing RA, Price HJ, Stelly DM (1995) Fluorescent in situ hybridization of a bacterial artificial chromosome. Genome 38:646–651
Kohel RJ, Stelly DM, Yu J (2002) Tests of six cotton (Gossypium hirsutum L.) mutants for association with aneuploids. J Hered 93:130–132
Lacape JM, Nguyen TB, Courtois B, Belot JL, Giband M, Gourlot JP, Gawryziak G, Roques S, Hau B (2005) QTL analysis of cotton fiber quality using multiple Gossypium hirsutum × Gossypium barbadense backcross generations. Crop Sci 45:123–140
Lacape JM, Nguyen TB, Thibivilliers S, Bojinov B, Courtois B, Cantrell RG, Burr B, Hau B (2003) A combined RFLP-SSR-AFLP map of tetraploid cotton based on a Gossypium hirsutum × Gossypium barbadense backcross population. Genome 46:612–626
Lichtenzveig J, Scheuring C, Dodge J, Abbo S, Zhang HB (2005) Construction of BAC and BIBAC libraries and their applications for generation of SSR markers for genome analysis of chickpea, Cicer arietinum L. Theor Appl Genet 110:492–510
Liu S, Saha S, Stelly DM, Burr B, Cantrell RG (2000) Chromosomal assignment of microsatellite loci in cotton. J Hered 91:326–332
Nguyen TB, Giband M, Brottier P, Risterucci AM, Lacape JM (2004) Wide coverage of the tetraploid cotton genome using newly developed microsatellite markers. Theor Appl Genet 109:167–176
Park YH, Alabady MS, Ulloa M, Sickler B, Wilkins TA, Yu J, Stelly DM, Kohel RJ, El-Shihy OM, Cantrell RG (2005) Genetic mapping of new cotton fiber loci using EST-derived microsatellites in an interspecific recombinant inbred line cotton population. Mol Gen Genomics 274:428–441
Rong J, Abbey C, Bowers JE, Brubaker CL, Chang C, Chee PW, Delmonte TA, Ding X, Garza JJ, Marler BS, Park C, Pierce GJ, Rainey KM, Rastogi VK, Schulze SR, Trolinder NL, Wendel JF, Wilkins TA, Williams-Coplin TD, Wing RA, Wright RJ, Zhao X, Zhu L, Paterson AH (2004) A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics 166:389–417
Saha S, Stelly DM (1994) Chromosomal location of phosphoglucomutase7 locus in Gossypium hirsutum. J Hered 85:35–40
Song X, Wang K, Guo W, Zhang J, Zhang T (2005) A comparison of genetic maps constructed from haploid and BC1 mapping populations from the same crossing between Gossypium hirsutum L. and Gossypium barbadense L. Genome 48:378–390
Ulloa M, Saha S, Jenkins JN, Meredith WR, McCarty JC, Stelly DM (2005) Chromosomal assignment of RFLP linkage groups harboring important QTLs on an intraspecific cotton (Gossypium hirsutum L.) joinmap. J Hered 96:132–144
Wang K, Guo W, Zhang T (2007) Development of one set of chromosome-specific microsatellite-containing BACs and their physical mapping in Gossypium hirsutum L. Theor Appl Genet 115:675–682
Wang K, Song X, Han Z, Guo W, Yu JZ, Sun J, Pan J, Kohel RJ, Zhang T (2006) Complete assignment of the chromosomes of Gossypium hirsutum L. by translocation and fluorescence in situ hybridization mapping. Theor Appl Genet 113:73–80
Wendel JF, Albert VA (1992) Phylogenetics of the cotton genus (Gossypium): character-state weighted parsimony analysis of chloroplast-DNA restriction site data and its systematic and biogeographic implications. Sys Bot 17:115–143
Werner-Fraczek JE, Close TJ (1998) Genetic studies of Triticeae dehydrins: assignment of seed proteins and a regulatory factor to map positions. Theor Appl Genet 97:220–226
Wu C, Sun S, Nimmakayala P, Santos F, Meksem K, Springman R, Ding K, Lightfoot DA, Zhang HB (2004) A BAC- and BIBAC-based physical map of the soybean genome. Genome Res 14:319–326
Yin J, Guo W, Yang L, Liu L, Zhang T (2006) Physical mapping of the Rf 1 fertility-restoring gene to a 100 kb region in cotton. Theor Appl Genet 112:1318–1325
Yu JZ, Kohel RJ, Dong J (2002a) Development of integrative SSR markers from TM-1 BACs. Proceeding of Beltwide cotton improvement conference, Atlanta, GA, January 8–12, 2002. CD-Rom Published by National Cotton Council of America
Yu JZ, Kohel RJ, Zhang HB, Dong J, Sun S, Steele NL (2002b) Toward development of a whole-genome, BAC/BIBAC-based integrated physical/genetic map of the cotton genome using the Upland genetic standard TM-1: BAC and BIBAC library construction, SSR marker development, and physical/genetic map integration. The third international cotton genome initiative workshop, Nanjing, China, June 3–6, 2002. Cotton Sci 14(S):32
Yu JZ, Kohel RJ, Zhang HB, Stelly D M, Xu Z, Dong J, Covaleda L, Lee MK, Cui P, Lazo GR, Gupta P, Ding K (2004a) Toward an integrative physical and genetic map of the cultivated allotetraploid cotton genome. Proceedings of the 12th International Conference on Plant and Animal Genome, San Diego, CA, January 10–16, 2004. P. No. W147 http://www.intlpag.org/12/abstracts/W32_PAG12_147.html. Cited 25 Oct 2007
Yu JZ, Kohel RJ, Zhang HB, Xu Z, Dong J (2004b) Integrated physical mapping of the cotton genome. Proceedings of the fourth International Cotton Genome Initiative (ICGI) Workshop, Hyderabad, India, October 10–13, 2004. Books of Abstracts, Published by CIRCOT, Mumbai, India. P. 2
Zhang P, Li W, John F, Bernd F, Bikram SG (2004) BAC-FISH in wheat identifies chromosome landmarkers consisting of different types of transposable elements. Chromosoma 112:288–299
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclaimer: Mention of trademark, proprietary product, or vendor does not constitute a guarantee or warranty of the product by USDA, ARS and does not imply its approval to the exclusion of other products or vendors that may also be suitable.
The U.S. Government’s right to retain a non-exclusive, royalty-free license in and to any copyright is acknowledged.
Rights and permissions
About this article
Cite this article
Guo, Y., Saha, S., Yu, J.Z. et al. BAC-derived SSR markers chromosome locations in cotton. Euphytica 161, 361–370 (2008). https://doi.org/10.1007/s10681-007-9585-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-007-9585-1