Abstract
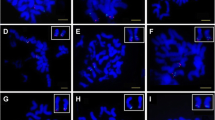
Fluorescence in situ hybridization (FISH), using bacterial artificial chromosome (BAC) clone as probe, is a reliable cytological technique for chromosome identification. It has been used in many plants, especially in those containing numerous small chromosomes. We previously developed eight chromosome-specific BAC clones from tetraploid cotton, which were used as excellent cytological markers for chromosomes identification. Here, we isolated the other chromosome-specific BAC clones to make a complete set for the identification of all 26 chromosome-pairs by this technology in tetraploid cotton (Gossypium hirsutum L.). This set of BAC markers was demonstrated to be useful to assign each chromosome to a genetic linkage group unambiguously. In addition, these BAC clones also served as convenient and reliable landmarks for establishing physical linkage with unknown targeted sequences. Moreover, one BAC containing an EST, with high sequence similarity to a G. hirsutum ethylene-responsive element-binding factor was located physically on the long arm of chromosome A7 with the help of a chromosome-A7-specific BAC FISH marker. Comparative analysis of physical marker positions in the chromosomes by BAC-FISH and genetic linkage maps demonstrated that most of the 26 BAC clones were localized close to or at the ends of their respective chromosomes, and indicated that the recombination active regions of cotton chromosomes are primarily located in the distal regions. This technology also enables us to make associations between chromosomes and their genetic linkage groups and re-assign each chromosome according to the corresponding genetic linkage group. This BAC clones and BAC-FISH technology will be useful for us to evaluate grossly the degree to which a linkage map provides adequate coverage for developing a saturated genetic map, and provides a powerful resource for cotton genomic researches.



Similar content being viewed by others
References
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–218
Baker RJ, Longmire JL, Van Den Bussche RA (1995) Organization of repetitive elements in the upland cotton genome (Gossypium hirsutum). J Hered 86:178–185
Brown MS (1980) The identification of the chromosomes of Gossypium hirsutum L. by means of translocations. J Hered 71:266–274
Cheng ZK, Yang XM, Yu HX, Gu MH (1998) A study of the number of SAT-chromosome in rice. Acta Genet Sin 25:225–231
Cheng ZK, Buell CR, Wing RA, Gu M, Jiang J (2001) Toward a cytological characterization of the rice genome. Genome Res 11:2133–2141
De Donato M, Gallagher DS, Davis SK, Ji Y, Burzlaff JD, Stelly DM, Womack JE, Taylor JF (1999) Physical assignment of microsatellite-containing BACs to bovine chromosomes. Cytogenet Cell Genet 87:59–61
Dong F, Song J, Naess SK, Helgeson JP, Gebhardt C, Jiang J (2000) Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor Appl Genet 101:1001–1007
Guo WZ, Cai CP, Wang CB, Han ZG, Song XL, Wang K, Niu XW, Wang C, Lu KY, Shi B, Zhang TZ (2007) A microsatellite-based, gene-rich linkage map reveals genome structure, function, and evolution in Gossypium. Genetics (in press)
Han ZG, Wang CB, Song XL, Guo WZ, Gou JY, Li CH, Chen XY, Zhang TZ (2006) Characteristics, development and mapping of G.hirsutum derived-EST-SSRs in allotetraploid cotton. Theor Appl Genet 112:430–439
Ji YF, Raska DA, McKnight TD, Islam-Faridi NM, Crane CF, Zwick MS, Hanson RE, Price HJ, Stelly DM (1997) Use of meiotic fluorescence in situ hybridization for identification of a new monosome in Gossypium hirsutum L. Genome 40:34–40
Jiang J, Gill BS (1994) Nonisotopic in situ hybridization and plant genome mapping: the first ten years. Genome 37:717–725
Jiang J, Gill BS, Wang GL, Ronald PC, Ward DC (1995) Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci USA 92:4487–4491
Kim JS, Childs KL, Islam-Faridi MN, Menz MA, Klein RR, Klein PE, Price HJ, Mullet JE, Stelly DM (2002) Integrated karyotyping of sorghum by in situ hybridization of landed BACs. Genome 45:402–412
Kohel RJ (1973) Genentic nomenclature in cotton. J Hered 64:291–295
Lacape JM, Nguyen TB, Thibivilliers S, Bojinov B, Courtois B, Cantrell RG, Burr B, Hau B (2003) A combined RFLP–SSR–AFLP map of tetraploid cotton based on a Gossypium hirsutum × Gossypium barbadense backcross population. Genome 46:612–626
Lee JA (1984) Cotton as a world crop. In: Kohel RJ, Lewis CL (eds) Cotton. Agronomy Monograph, No. 24. Crop Science Society of America, Madison, Wisconsin, pp 1–25
Levy AA, Feldman M (2002) The impact of polyploidy on grass genome evolution. Plant Physiol 130:1587–1593
McClintock B (1929) Chromosome morphology in Zea mays. Science 69:629
Menzel MY (1954) A cytological method for genome analysis in Gossypium. Genetics 40:214–223
Nandi HK (1936) The chromosome morphology, second association and origin of cultivated rice. J Genet 33:315–316
Okamoto M (1962) Identification of the chromosomes of common wheat belonging to the A and B genome. Can I Genet Cytol 4:31–37
Rayburn AL, Gill BS (1985) Use of biotin-labeled probes to map specific DNA sequences of wheat chromosomes. J Hered 76:78–81
Rick CM, Barton DW (1954) Cytological and genetical identification of primary trisomics of the tomato. Genetics 39:640–666
Sears ER (1969) Wheat cytogenetics. Ann Rev Genet 3:451–468
Wang K, Song XL, Han ZG, Guo WZ, Yu JZ, Sun J, Pan JJ, Kohel RJ, Zhang TZ (2006) Complete assignment of the chromosomes of Gossypium hirsutum L. by translocation and fluorescence in situ hybridization mapping. Theor Appl Genet 113:73–80
Wang K, Zhang YJ, Zhang TZ (2005) A high throughput approach for cotton BAC-DNA isolation. Cotton Sci 17:125–126
Wendel JF (1989) New world cottons contain Old World cytoplasm. Proc Natl Acad Sci USA 86:4132–4136
Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249
Wendel JF, Cronn RC (2002) Polyploidy and the evolutionary history of cotton. Adv Agron 78:139–186
Yin JM, Guo WZ, Yang LM, Liu LW, Zhang TZ (2006) Physical mapping of the Rf 1 fertility-restoring gene to a 100 kb region in cotton. Theor Appl Genet 112:1318–1325
Yu HG, Liang H, Kofoid KD (1991) Analysis of C-banding chromosome patterns of sorghum. Crop Sci 31:1524–1527
Zhang P, Li WL, Fellers J, Friebe B, Gill BS (2004) BAC-FISH in wheat identifies chromosome landmarks consisting of different types of transposable elements. Chromosoma 112:288–299
Acknowledgments
This work was financially supported in part by grants from the program of Changjiang Scholars and Innovative Research Team in University of MOE, China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. S. Heslop-Harrison.
Rights and permissions
About this article
Cite this article
Wang, K., Guo, W. & Zhang, T. Development of one set of chromosome-specific microsatellite-containing BACs and their physical mapping in Gossypium hirsutum L.. Theor Appl Genet 115, 675–682 (2007). https://doi.org/10.1007/s00122-007-0598-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-007-0598-x