Abstract
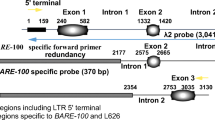
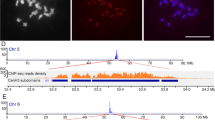
Although a centromeric DNA fragment of tobacco (Nicotiana tabacum), Nt2-7, has been reported, the overall structure of the centromeres remains unknown. To characterize the centromeric DNA sequences, we conducted a chromatin immunoprecipitation assay using anti-NtCENH3 antibody and chromatins isolated from two ancestral diploid species (Nicotiana sylvestris and Nicotiana tomentosiformis) of N. tabacum and isolated a 178-pb fragment, Nto1 from N. tomentosiformis, as a novel centromeric DNA. Fluorescence in situ hybridization (FISH) showed that Nto1 localizes on 24 out of 48 chromosomes in some cells of a BY-2 cell line. To identify the origins of the Nt2-7 and Nto1, a tobacco bacterial artificial chromosome (BAC) library was constructed from N. tabacum, and then screened by polymerase chain reaction (PCR) with primer sets designed from the Nt2-7 and Not1 DNA sequences. Twelve BAC clones were found to localize on the centromeric regions by FISH. We selected three BAC clones for sequencing and identified two centromeric retrotransposons, NtCR and NtoCR, the DNA sequences of which are similar to that of Nt2-7 and Nto1, respectively. Quantitative PCR analysis using coprecipitated DNA with anti-NtCENH3 clearly showed coexistence of NtCENH3 with both retrotransposons. These results indicate the possibility that these two retrotransposons act as centromeric DNA sequences in tobacco. NtoCR was found to be specific to N. tomentosiformis and T genome of N. tabacum, and a NtCR-like centromeric retrotransposon (TGRIV) exists in tomato. This specificity suggests that the times of amplification of these centromeric retrotransposons were different.








Similar content being viewed by others
Abbreviations
- ANOVA:
-
Analysis of variance
- Av:
-
Average
- BAC:
-
Bacterial artificial chromosome
- BLAST:
-
The basic local alignment search tool
- BLASTN:
-
BLAST for nucleotide
- CENH3:
-
Centromere-specific histone H3
- CentC:
-
Centromeric repeat type C in maize
- CentO:
-
Centromeric tandem repeat in Oryza sativa
- CHEF:
-
Clamped homogeneous electric fields
- ChIP:
-
Chromatin immunoprecipitation
- CR:
-
Centromeric retrotransposon
- CRM:
-
Centromeric retrotransposon in maize
- CRR:
-
Centromeric retrotransposon in rice
- CRW:
-
Centromeric retrotransposon in wheat
- DAPI:
-
4,6-Diamino-2-phenylindole
- DIG:
-
Digoxigenin
- EDTA:
-
Ethylendiaminetetraacetic acid
- FIGE:
-
Field inversion gel electrophoresis
- FISH:
-
Fluorescence in situ hybridization
- FITC:
-
Fluorescein isothiocianate
- IPTG:
-
Isopropylthio-β-d-galactoside
- LB medium:
-
Luria–Bertani medium
- LTR:
-
Long terminal repeat
- MES:
-
2-(N-morpholino) ethanesulfonic acid
- MYA:
-
Million years ago
- NtCENH3:
-
Nicotiana tabacum centromere-specific histone H3
- NtCR:
-
Nicotiana tabacum centromeric retrotransposon
- NtCR-NA:
-
Nicotiana tabacum centromeric retrotransposon non-autonomous type
- NtoCR:
-
Nicotiana tomentosiformis centromeric retrotransposon
- Tukey HSD:
-
Tukey’s honestly significant difference
- Pel fraction:
-
Bound fractions in ChIP
- PMSF:
-
Phenylmethylsulfonyl fluoride
- qPCR:
-
Real-time quantitative PCR
- SE:
-
Standard error
- SDS:
-
Sodium dodecyl sulfate
- STE lysis buffer:
-
SDS Tris and EDTA lysis buffer
- TBS:
-
Tris-buffered saline
- TGRIV:
-
Tomato genomic repeat IV
- Tris:
-
Tris (hydroxymethyl) aminomethane
- X-gal:
-
5-Bromo-4-chloro-3-indolyl-β-d-galactoside
References
Aragon-Alcaide L, Miller T, Schwarzacher T, Reader S, Moore G (1996) A cereal centromeric sequence. Chromosoma 105:261–268
Chang SB, Yang TJ, Datema E, van Vugt J, Vosman B, Kuipers A, Meznikova M, Szinay D, Lankhorst RK, Jacobsen E, de Jong H (2008) FISH mapping and molecular organization of the major repetitive sequences of tomato. Chromosome Res 16:919–933
Cheng Z, Dong F, Langdon T, Ouyang S, Buell CR, Gu M, Blattner FR, Jiang J (2002) Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14:1691–1704
Choo KHA (1997) The centromere. Oxford University Press, Oxford
Clarke L, Carbon J (1983) Genomic substitutions of centromeres in Saccharomyces cerevisiae. Nature 305:23–28
Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S, Leitch AR (2005) Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae). New Phytol 168:241–252
Devos KM, Brown JK, Bennetzen JL (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res 12:1075–1079
Hirochika H, Otsuki H, Yoshikawa M, Otsuki Y, Sugimoto K, Takeda S (1996) Autonomous transposition of the tobacco retrotransposon Tto1 in rice. Plant Cell 8:725–734
Hosouchi T, Kumekawa N, Tsuruoka H, Kotani H (2002) Physical map-based sizes of the centromeric regions of Arabidopsis thaliana chromosomes 1, 2, and 3. DNA Res 9:117–121
Houben A, Schroeder-Reiter E, Nagaki K, Nasuda S, Wanner G, Murata M, Endo TR (2007) CENH3 interacts with the centromeric retrotransposon cereba and GC-rich satellites and locates to centromeric substructures in barley. Chromosoma 116:275–283
Hudakova S, Michalek W, Presting GG, ten Hoopen R, dos Santos K, Jasencakova Z, Schubert I (2001) Sequence organization of barley centromeres. Nucleic Acids Res 29:5029–5035
International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Jiang J, Nasuda S, Dong F, Scherrer CW, Woo S, Wing RA, Gill BS, Ward DC (1996) A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc Natl Acad Sci USA 93:14210–14213
Jiang J, Birchler JA, Parrott WA, Dawe RK (2003) A molecular view of plant centromeres. Trends Plant Sci 8:570–575
Jin W, Melo JR, Nagaki K, Talbert PB, Henikoff S, Dawe RK, Jiang J (2004) Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell 16:571–581
Kato K, Matsumoto T, Koiwai A, Mizusaki S, Nishida K, Noguchi M, Tamaki E (1972) Liquid suspension culture of tobacco cells. In: Terui G (ed) Fermentation technology today. Osaka, Society of Fermentation Technology of Japan, pp 689–695
Kim U-J, Birren BW, Slepak T, Mancino V, Boysen C, Kang H-L, Simon MI, Shizuya H (1996) Construction and characterization of a human bacterial artificial chromosome library. Genomics 34:213–218
Kumekawa N, Hosouchi T, Tsuruoka H, Kotani H (2000) The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 5. DNA Res 7:315–321
Kumekawa N, Hosouchi T, Tsuruoka H, Kotani H (2001) The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 4. DNA Res 8:285–290
Lee H, Zhang W, Langdon T, Jin W, Yan H, Cheng Z, Jiang J (2005) Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci USA 102:11793–11798
Lim KY, Skalicka K, Koukalova B, Volkov RA, Matyasek R, Hemleben V, Leitch AR, Kovarik A (2004) Dynamic changes in the distribution of a satellite homologous to intergenic 26-18S rDNA spacer in the evolution of Nicotiana. Genetics 166:1935–1946
Liu Z, Yue W, Li D, Wang RR, Kong X, Lu K, Wang G, Dong Y, Jin W, Zhang X (2008) Structure and dynamics of retrotransposons at wheat centromeres and pericentromeres. Chromosoma 117:445–456
Malik HS, Henikoff S (2002) Conflict begets complexity: the evolution of centromeres. Curr Opin Genet Dev 12:711–718
Moscone EA, Matzke MA, Matzke AJ (1996) The use of combined FISH/GISH in conjunction with DAPI counterstaining to identify chromosomes containing transgene inserts in amphidiploid tobacco. Chromosoma 105:231–236
Murata M (2002) Telomeres and centromeres in plants. Curr Genet 3:527–538
Nagaki K, Murata M (2005) Characterization of CENH3 and centromere-associated DNA sequences in sugarcane. Chromosome Res 13:195–203
Nagaki K, Talbert PB, Zhong CX, Dawe RK, Henikoff S, Jiang J (2003a) Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163:1221–1225
Nagaki K, Song J, Stuper RM, Parokonny AS, Yuan Q, Ouyang S, Liu J, Hsiao J, Jones KM, Dawe RK, Buell CR, Jiang J (2003b) Molecular and cytological analyses of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics 163:759–770
Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36:138–145
Nagaki K, Neumann P, Zhang D, Ouyang S, Buell CR, Cheng Z, Jiang J (2005) Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol Biol Evol 22:845–855
Nagaki K, Kashihara K, Murata M (2009a) A centromeric DNA sequence colocalized with a centromere-specific histone H3 in tobacco. Chromosoma 118:249–257
Nagaki K, Walling J, Hirsch C, Jiang J, Murata M (2009b) Structure and evolution of plant centromeres. In: Ugarkovic D (ed) Centromere, progress in molecular and subcellular biology. Springer, Berlin, pp 153–179
Pelissier T, Tutois S, Deragon JM, Tourmente S, Genestier S, Picard G (1995) Athila, a new retroelement from Arabidopsis thaliana. Plant Mol Biol 29:441–452
Petit M, Lim KY, Julio E, Poncet C, Borne FDD, Kovarik A, Leitch AR, Grandbastien M-A, Mhiri C (2007) Differential impact of retrotransposon populations on the genome of allotetraploid tobacco (Nicotiana tabacum). Mol Genet Genomics 278:1–15
Suzuki G, Watanabe M, Toriyama K, Isogai A, Hinata K (1997) Direct cloning of the Brassica S locus by using a P1-derived artificial chromosome (PAC) vector. Gene 199:133–137
Tek AL, Kashihara K, Murata M, Nagaki K (2010) Functional centromeres in soybean include two distinct tandem repeats and a retrotransposon. Chromosome Res 18:337–347
The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Wolfgruber TK, Sharma A, Schneider KL, Albert PS, Koo D-H, Shi J, Gao Z, Han F, Lee H, Xu R, Allison J, Birchler JA, Jiang J, Dawe RK, Presting GG (2009) Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic loci shaped primarily by retrotransposons. PLoS Genet 5:e1000743
Wu F, Tanksley SD (2010) Chromosomal evolution in the plant family Solanaceae. BMC Genomics 11:182
Yan H, Ito H, Nobuta K, Ouyang S, Jin W, Tian S, Lu C, Venu RC, Wang GL, Green PJ, Wing RA, Buell CR, Meyers BC, Jiang J (2006) Genomic and genetic characterization of rice Cen3 reveals extensive transcription and evolutionary implications of a complex centromere. Plant Cell 18:2123–2133
Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, Nagaki K, Birchler JA, Jiang J, Dawe RK (2002) Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14:2825–2836
Acknowledgments
This work was supported by the Program for the Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN) and a grant from the Inamori Foundation (#200835).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Pat Heslop-Harrison.
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Figure S1
Multi-color FISH on SR1 chromosomes. DAPI-stained N. tabacum SR1 chromosomes (b), NtBAC#1 (c), #2 (d), #9 (e), and #10 (f) FISH signals, and merged image of b (white), c (red), d (yellow), e (green), and f (blue) (a). Scale bar, 10 μm (JPEG 48 kb)
Figure S2
Immunostaining and FISH. DAPI-stained N. tabacum chromosomes (b, f and j), immunosignals of anti-NtCENH3 antibody (c, g and k), and NtBAC#2 (d), #9 (h), and #10 (l) FISH signals and merged image of b–d (a), f–h (e), and j–l (i). Scale bar, 10 μm (JPEG 95 kb)
Table S1
Primers used in this study (DOC 115 kb)
Table S2
Tobacco BAC clones showing centromeric FISH signals (DOC 45 kb)
Rights and permissions
About this article
Cite this article
Nagaki, K., Shibata, F., Suzuki, G. et al. Coexistence of NtCENH3 and two retrotransposons in tobacco centromeres. Chromosome Res 19, 591–605 (2011). https://doi.org/10.1007/s10577-011-9219-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-011-9219-2