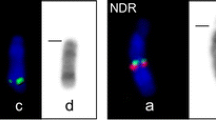
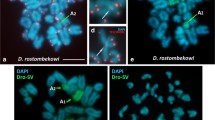
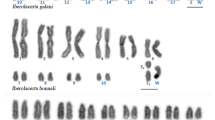
Abstract
The four-horned antelope, Tetracerus quadricornis, is a karyotypic novelty in Bovidae since chromosomal evolution in this species is driven by tandem fusions in contradiction to the overwhelming influence of Robertsonian fusions in other species within the family. Using a combination of differential staining and molecular cytogenetic techniques, we provide the first description of the species’ karyotype, draw phylogenetic inferences from the cytogenetic data and discuss possible mechanisms underlying the formation of the tandem fusions in this species. We show (a) that pairs 1–6 of Tetracerus correspond to a combination of Bos taurus orthologous chromosomes that are tandemly fused head to tail, (b) the presence of interstitial centromeric satellite DNA at the junctions of orthologous blocks defined by the cross-species painting data and (c) that in some instances, residual telomeric sequences persist at these sites. We conclude that the attendant result of each fusion is an enlarged acrocentric fusion element comprising a single functional centromere and two terminal telomeres that, collectively, led to a reduction of the 2n = 58 bovid ancestral acrocentric chromosomal complement to the 2n = 38 detected in the four-horned antelope.





Similar content being viewed by others
Abbreviations
- BTA:
-
Bos taurus
- DOP-PCR:
-
Degenerate oligonucleotide primed-PCR
- DAPI:
-
4′,6-diamidino-2-phenylindole
- FISH:
-
fluorescence in situ hybridisation
- NORs:
-
nucleolar organiser regions
- pter:
-
terminus of p arm
- q-dist:
-
distal q arm
- Rb fusions:
-
Robertsonian fusions
- cenDNA:
-
centromeric DNA
- TQU:
-
Tetracerus quadricornis
References
Adega F, Gueddes-Pinto H, Chaves R (2009) Satellite DNA in the karyotype evolution of domestic animals—clinical considerations. Cytogenet Genome Res 126:12–20
Bongso TA, Hilmi M (1982) Chromosome banding homologies of a tandem fusion in river, swamp and crossbred buffalo (Bubalus bubalis). Can J Genet Cytol 24:667–673
Buckland R, Evans H (1978) Cytogenetic aspects of phylogeny in the Bovidae. I. G-banding. Cytogenet Cell Genet 21:42–63
Chi JX, Huang L, Nie W et al (2005) Defining the orientation of the tandem fusions that occurred during the evolution of Indian muntjac chromosomes by BAC mapping. Chromosoma 114:167–172
Di Berardino D, Iannuzzi L (1981) Chromosome banding homologies in Swamp and Murrah buffalo. J Heredity 72:183–188
Elder FFB (1980) Tandem fusion, centric fusion, and chromosomal evolution in the cotton rats, genus Sigmodon. Cytogenet Cell Genet 26:199–210
Gallagher D, Womack J (1992) Chromosome conservation in the Bovidae. J Heredity 83:287–298
Gallagher DS Jr, Davis SK, De Donato M et al (1998) A karyotypic analysis of nilgai, Boselaphus tragocamelus (Artiodactyla: Bovidae). Chromosome Res 6:505–513
Gallagher DS Jr, Davis SK, De Donato M et al (1999) A molecular cytogenetic analysis of the tribe Bovini (Artiodactyla: Bovidae: Bovinae) with an emphasis on sex chromosome morphology and NOR distribution. Chromosome Res 7:481–492
Genome 10K Community of Scientists (2009) Genome 10K: a proposal to obtain whole-genome sequence for 10000 vertebrate species. J Hered 100:659–674
Goodpasture C, Bloom SE (1975) Visualization of nucleolar organizer in mammalian chromosomes using silver staining. Chromosoma 53:37–50
Hartmann N, Scherthan H (2004) Characterization of ancestral chromosome fusion points in the Indian muntjac deer. Chromosoma 112:213–220
Hassanane MS, Chaudhary R, Chowdhary BP (1998) Microdissected bovine X chromosome segment delineates homoeologous chromosomal regions in sheep, goat and buffalo. Chromosome Res 6:213–217
Hassanin A, Douzery E (1999a) Evolutionary affinities of the enigmatic saola (Pseudoryx nghetinhensis) in the context of the molecular phylogeny of Bovidae. Proc Biol Sci 266:893–900
Hassanin A, Douzery E (1999b) The tribal radiation of the family Bovidae (Artiodactyla) and the evolution of the mitochondrial cytochrome b gene. Mol Phylogenet Evol 13:227–243
Hassanin A, Ropiquet A (2004) Molecular phylogeny of the tribe Bovini (Bovidae, Bovinae) and the taxonomic status of the Kouprey, Bos sauveli Urbain, 1937. Mol Phylogenet Evol 33:896–907
Hassanin A, Ropiquet A (2007) Resolving a zoological mystery: the Kouprey is a real species. Proc Biol Sci 274:2849–2855
Hassanin A, Ropiquet A, Couloux A et al (2009) Evolution of the mitochondrial genome in mammals living at high altitude: new insights from a study of the tribe Caprini (Bovidae, Antilopinae). Mol Phylogenet Evol 68:293–310
Huang L, Wang J, Nie W et al (2006a) Tandem fusions in karyotype evolution of Muntiacus: evidence from M. feae and M. gongshanensis. Chromosome Res 14:637–647
Huang L, Chi J, Wang J et al (2006b) High-density comparative mapping in the black muntjac (Muntiacus crinifrons): molecular cytogenetic dissection of the origin of MCR 1p + 4 in the X1X2Y1Y2Y3 sex chromosome system. Genomics 87:608–615
Iannuzzi L, Di Meo GP (1995) Chromosomal evolution in bovids: a comparison of cattle, sheep and goat G- and R-banded chromosomes and cytogenetic divergences among cattle, goat and river buffalo sex chromosomes. Chromosome Res 3:291–299
ISCNDB, Cribiu EP, Di Berardino D, Di Meo GP et al (2001) International System for Chromosome Nomenclature of Domestic Bovids (ISCNDB 2000). Cytogenet Cell Genet 92:283–299
Krishna YC, Clyne PJ, Krishnaswamy J et al (2009) Distributional and ecological review of the four horned antelope, Tetracerus quadricornis. Mammalia 73:1–6
Kubickova S, Cernohorska H, Musilova P et al (2002) The use of laser microdissection for the preparation of chromosome-specific painting probes in farm animals. Chromosome Res 10:571–577
Kumamoto AT, Kingswood SC, Wouter H (1994) Chromosomal divergence in allopatric populations of kirk's dik-dik, Madoqua kirki (Artiodactyla, Bovidae). J Mammal 75:357–364
Maddison WP, Maddison DR (2003) MacClade: analysis of phylogeny and character evolution, version 4.06. Sinauer, Sunderland
Nguyen TT, Aniskin VM, Gerbault-Seureau M et al (2008) Phylogenetic position of the saola (Pseudoryx nghetinhensis) inferred from cytogenetic analysis of eleven species of Bovidae. Cytogenet Genome Res 122:41–54
Nijman IJ, van Boxtel DCJ, van Cann LM et al (2008) Phylogeny of Y chromosomes from bovine species. Cladistics 24:723–726
O’Brien SJ, Menninger JC, Nash WG (eds) (2006) Atlas of mammalian chromosomes. Wiley, Hoboken
Robinson TJ, Harrison WR, Ponce de Leon A et al (1998) A molecular cytogenetic analysis of X chromosome repatterning in the Bovidae: transpositions, inversions and phylogenetic inference. Cytogenet Cell Genet 80:179–184
Ropiquet A, Hassanin A (2005) Molecular evidence for the polyphyly of the genus Hemitragus (Mammalia, Bovidae). Mol Phylogenet Evol 36:154–168
Ropiquet A, Hassanin A (2006) Hybrid origin of the Pliocene ancestor of wild goats. Mol Phylogenet Evol 41:395–404
Ropiquet A, Gerbault-Seureau M, Deuve JL et al (2008) Chromosome evolution in the subtribe Bovina (Mammalia, Bovidae): the karyotype of the Cambodian banteng (Bos javanicus birmanicus) suggests that Robertsonian translocations are related to interspecific hybridization. Chromosome Res 16:1107–1118
Ropiquet A, Li B, Hassanin A (2009) SuperTRI: a new approach based on branch support analyses of multiple independent data sets for assessing reliability of phylogenetic inferences. C R Biol 332:832–847
Rubes J, Kubickova S, Pagacova E et al (2008) Phylogenomic study of spiral-horned antelope by cross-species chromosome painting. Chromosome Res 16:935–947
Scherthan H (1990) The localization of the repetitive telomeric sequence (TTAGGG) n in two muntjac species and implications for their karyotypic evolution. Cytogenet Cell Genet 53:115–117
Seabright M (1971) A rapid banding technique for human chromosomes. Lancet 2:971–972
Solounias N (1990) A new hypothesis uniting Boselaphus and Tetracerus with the Miocene Boselaphini (Mammalia, Bovidae) based on horn morphology. Annales Musei Goulandris 8:425–439
Sumner AT (1990) Chromosome banding. Unwin Hyman, London
Swier VJ, Bradley RD, Rens W et al (2009) Patterns of chromosomal evolution in Sigmodon, evidence from whole chromosome paints. Cytogenet Cell Genet 125:54–66
Tanaka K, Matsuda Y, Masangkay JS, Solis CD, Anunciado RVP, Namikawa T (1999) Characterization and chromosomal distribution of satellite DNA sequences of the water buffalo (Bubalus bubalis). Heredity 90:418–422
Willows-Munro S, Robinson TJ, Matthee CA (2005) Utility of nuclear DNA intron markers at lower taxonomic levels: phylogenetic resolution among nine Tragelaphus spp. Mol Phylogenet Evol 35:624–636
Wilson DE, Reeder DM (2005) Mammal species of the world. Johns Hopkins University Press, Baltimore
White MJD (1978) Modes of speciation. Freeman, San Francisco
Wurster D, Benirschke K (1968) Chromosome studies in the superfamily Bovoidea. Chromosoma 25:152–171
Acknowledgements
Tetracerus skin biopsies were provided by Norin Chai (Ménagerie du Jardin des Plantes, MNHN). Financial support from the MNHN, CNRS, the PPF “Etat et structure phylogénétique de la biodiversité actuelle et fossile” (A.H and A.R) as well as the National Research Foundation of South Africa (TJR and AR) is gratefully acknowledged. V.R.I. was funded through research grants MZE 0002716202 and GA CR P506/10/0421.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible Editor: Herbert Macgregor.
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Table S1
Matrix of cytogenetic characters used in this study. Characters that are considered exclusive synapomorphies—red; homoplasies—green; autapomorphies—grey (DOC 1003 kb)
Rights and permissions
About this article
Cite this article
Ropiquet, A., Hassanin, A., Pagacova, E. et al. A paradox revealed: karyotype evolution in the four-horned antelope occurs by tandem fusion (Mammalia, Bovidae, Tetracerus quadricornis). Chromosome Res 18, 277–286 (2010). https://doi.org/10.1007/s10577-010-9115-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-010-9115-1