Abstract
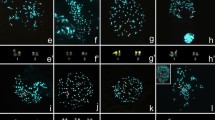
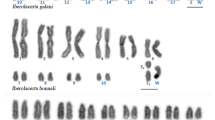
The muntjacs (Muntiacus, Cervidae) are famous for their rapid and radical karyotypic diversification via repeated tandem chromosome fusions, constituting a paradigm for the studies of karyotypic evolution. Of the five muntjac species with defined karyotypes, three species (i.e. Muntiacus reevesi, 2n = 46; M. m. vaginalis, 2n = 6/7; and M. crinifrons, 2n = 8/9) have so far been investigated by a combined approach of comparative chromosome banding, chromosome painting and BAC mapping. The results demonstrated that extensive centromere–telomere fusions and a few centric fusions are the chromosomal mechanisms underlying the karyotypic evolution of muntjacs. Here we have applied the same approach to two additional muntjac species with less well-characterized karyotypes, M. feae (2n = 14♂) and M. gongshanensis (2n = 8♀). High-resolution G-banded karyotypes for M. feae and M. gongshanensis are provided. The integrated analysis of hybridization results led to the establishment of a high-resolution comparative map between M. reevesi, M. feae, and M. gongshanensis, proving that all tandem fusions underpinning the karyotypic evolution of these two muntjac species are also centromere–telomere fusions. Furthermore, the results have improved our understanding of the karyotypic relationships of extant muntjac species and provided compelling cytogenetic evidence that supports the view that M. crinifrons, M. feae, and M. gongshanensis should each be treated as a distinct species.
Similar content being viewed by others
References
Amato G, Egan MG, Rabinowitz A (1999a) A new species of muntjac, Muntiacus putaoensis (Artiodactyla: Cervidae) from northern Myanmar. Anim Conserv 2: 1–7.
Amato G, Egan MG, Schaller GB et al. (1999b) Rediscovery of Roosevelt’s Barking Deer (Muntiacus Rooseveltorum). J Mammal 80: 639–643.
Amato G, Egan MG, Schaller GB (2000) Mitochondrial DNA variation in Muntjac: evidence for discovery, rediscovery, and phylogenetic relationships. In Vrba ES, Shaller GB, eds., Antelopes, Deer, and Relatives: Fossil Record, Behavioral Ecology, Systematics, and Conservation. New Haven: Yale University Press, pp. 285–295.
Benedum UMH, Neitzel H, Sperling K, Bogenberger J, Fittler F (1986) Organization and chromosomal distribution of a novel repetitive DNA component from Muntiacus muntjak vaginalis with a repeat length of more than 40 kb. Chromosoma 94: 267–272.
Bogenberger JM, Neumaier PS, Fittler F (1985) The muntjak satellite IA sequence is composed of 31-base-pair internal repeats that are highly homologous to the 31-base-pair subrepeats of the bovine satellite 1.715. Eur J Biochem 148: 55–59.
Bogenberger JM, Neumaier PS, Fittler F (1987) A highly repetitive DNA component common to all Cervidae: its organization and chromosomal distribution during evolution. Chromosoma 95: 154–161.
Chi J, Fu B, Nie W, Wang J, Graphodatsky A, Yang F (2005a) New insights into the karyotypic relationships of Chinese muntjac (Muntiacus reevesi), forest musk deer (Moschus berezovskii) and gayal (Bos frontalis). Cytogenet Genome Res 108: 310–316.
Chi J, Huang L, Nie W, Wang J, Su B, Yang F (2005b) Defining the orientation of the tandem fusions that occurred during the evolution of Indian muntjac chromosomes by BAC mapping. Chromosoma 114: 167–172.
Corbet GB, Hill JE (1992) The Mammals of the Indomalayan Region: A Systematic Review. Natural History Museum Publications. London: Oxford University Press, pp. 259–261.
Datta A, Pansa J, Madhusudan MD, Mishra C (2003) Discovery of the leaf deer Muntiacus putaoensis in Arunachal Pradesh: an addition to the large mammals of Indian. Current Sci 84: 454–458.
Elder FFB, Hsu TC (1988) Tandem fusions in the evolution of mammalian chromosomes. In Sandberg AA, ed., The Cytogenetics of Mammalian Autosomal Rearrangements. New York: Alan R. Liss, pp. 481–506.
Evans TD, Timmins RJ (1994) News from Laos. Oryx 29: 3–4.
Fontana F, Rubini M (1990) Chromosomal evolution in Cervidae. Biosystem 24: 157–174.
Frönicke F, Scherthan H (1997) Zoo-fluorescence in situ hybridization analysis of human and Indian muntjac karyotypes (Muntiacus muntjak vaginalis) reveals satellite DNA clusters at the margins of conserved syntenic segments. Chromosome Res 5: 251–261.
Frönicke L, Chowdhary BP, Scherthan H (1997) Segmental homology among cattle (Bos taurus), Indian muntjac (Muntiacus muntjak vaginalis), and Chinese muntjac (M. reevesi) karyotype. Cytogenet Cell Genet 77: 223–227.
Giao PM, Touc D, Dung VV et al. (1998) Description of Muntiacus truongsonensis, a new species of muntjac (Artiodactyla: Muntiacidae) from central Vietnam, and implications for conservation. Animal Conserv 1: 61–68.
Groves CP (1982) The species of muntjac (Genus Muntiacus) in Borneo: unrecognized sympatry in tropical deer. Zool Medelingen 56: 203–216.
Groves CP, Grubb P (1987) Relationships of living deer. In Wemmer CM, ed., Biology and Management of the Cervidae. Washington, DC: Smithsonian Institution, pp. 21–59.
Groves CP, Grubb P (1990) Muntiacidae. In Bubenik GA, Bubenik AB, eds., Horns, Pronghorns, and Antlers. New York: Springer-Verlag, pp. 134–168.
Hsu TC, Pathak S, Chen TR (1975) The possibility of latent centromeres and a proposed nomenclature system for total chromosome and whole arm translocations. Cytogenet Cell Genet 15: 41–49.
Huang L, Chi J, Nie W, Wang J, Yang F (2006a) Phylogenomics of several deer species revealed by comparative chromosome painting with Chinese muntjac paints. Genetica. DOI 10.1007/s10709-005-2449-5
Huang L, Chi J, Wang J, Nie W, Su W, Yang F (2006b) High-density comparative BAC mapping in the black muntjac (Muntiacus crinifrons): molecular cytogenetic dissection of the origin of MCR 1p+4 in the X1X2Y1Y2Y3 sex chromosome system. Genomics 87: 608–615.
Lee C, Lin CC (1996) Conservation of a 31bp bovine subrepeat in centromeric satellite DNA monomers of Cervus elaphus and other cervid species. Chromosome Res 4: 427–435.
Lee C, Sasi R, Lin CC (1993) Interstitial localization of telomeric DNA sequences in the Indian muntjac chromosomes: further evidence for tandem chromosome fusions in the karyotypic evolution of the Asian muntjacs. Cytogenet Cell Genet 63: 156–159.
Lee C, Ritchie DBC, Lin CC (1994) A tandem repetitive, centromeric DNA sequence from the Canadian woodland caribou (Rangifer tarandus caribou): its conservation and evolution in several deer species. Chromosome Res 2: 293–306.
Lee C, Court DR, Cho C, Haslett JL, Lin CC (1997) Higher-order organization of subrepeats and the evolution of cervid satellite I DNA. J Mol Evol 44: 327–335.
Li YC, Lee C, Hseu TH, Li SY, Lin CC (2000a) Direct visualization of the genomic distribution and organization of two cervid centromeric satellite DNA families. Cytogenet Cell Genet 89: 192–198.
Li YC, Lee C, Sanoudou D, Hseu TH, Li SY, Lin CC (2000b) Interstitial colocalization of two cervid satellite DNAs involved in the genesis of the Indian muntjac karyotype. Chromosome Res 8: 363–373.
Li YC, Lee C, Chang WS, Li SY, Lin CC (2002) Isolation and identification of a novel satellite DNA family highly conserved in several Cervidae species. Chromosoma 111: 176–183.
Lin CC, Sasi R, Fan YS, Chen ZQ (1991) New evidence for tandem chromosome fusions in the karyotypic evolution of Asian muntjac. Chromosoma 101: 19–24.
Lin CC, Chiang PY, Hsieh LJ, Liao SJ, Chao MC, Li YC (2004) Cloning, characterization and physical mapping of three cervid satellite DNA families in the genome of the Formosan muntjac (Muntiacus reevesi micrurus). Cytogenet Genome Res 105: 100–106.
Ma SL, Wang YX, Xu LH (1986) Taxonomic and phylogenetic studies on the genus Muntiacus. Acta Theriol Sin 6: 191–207.
Ma SL, Wang YX, Shi LM (1990) A new species of the genus Muntiacus from Yunnan, China. Zool Res 11: 47–53.
Nowak RM (1999) Walker’s Mammals of the World. Baltimore: Johns Hopkins University Press, pp. 1051–1135.
Schaller GB, Vrba ES (1996) Description of the giant muntjac (Megamuntiacus vaquangensis) in Laos. J Mammal 77: 675–683.
Scherthan H (1990) Localization of the repetitive telomeric sequence (TTAGGG)n in two muntjac species and implications for their karyotypic evolution. Cytogenet Cell Genet 53: 115–117.
Scherthan H (1995) Chromosome evolution in muntjac revealed by centromere, telomere and whole chromosome paint probes. Kew Chromosome Conference IV: 267–280.
Seabright M (1972) The use of proteolytic enzymes for the mapping of structural rearrangements in the chromosomes of man. Chromosoma 36: 204–210.
Sheng HL (1992) The Deer in China. Shanghai: East China Normal University Press.
Sheng HL (1998) Black muntjac (Muntiacus crinifrons), Fea’s muntjac (Muntiacus feae). In Wang S, ed., China Red Data Book of Endangered Animals (Mammalia). Beijing: Science Press, pp. 282–286.
Sheng HL, Lu HG (1990) A discussion regarding Fea’s muntjac, Muntiacus feae. J East China Norm Univ (Mamm Ecol Suppl), p. 121.
Shi LM (1983) Sex-linked chromosome polymorphism in black muntjac, Muntiacus crinifrons. In Swamina-Than MS, ed., Proceedings of the Fifth International Congress of Genetics, New Dehli, p. 153.
Shi LM, Ma CX (1988) A new karyotype of muntjac (Muntiacus sp.) from Gongshan county in China. Zool Res 9: 343–347.
Shi LM, Ye YY, Duan XS (1980) Comparative cytogenetic studies on the red muntjac, Chinese muntjac and their F1 hybrids. Cytogenet Cell Genet 26: 22–27.
Soma H, Kada H, Mitayoshi K et al. (1983) The chromosomes of Muntiacus feae. Cytogenet Cell Genet 35: 156–158.
Soma H, Kada H, Meckvichal C, Mahannop A (1987) Confirmation of the chromosome constitution of Fea’s muntjac, Muntiacus feae. Proc Jpn Acad B Phys Biol Sci 63: 253–256.
Tanomtong A, Chaveerach A, Phanjun G, Kaensa W, Khunsook S (2005) New records of chromosomal feature in Indian muntiac (Muntiacus muntjak) and Fea’s Muntjacs (M. feae) of Thailand. Cytologia 70: 71–77.
Touc D, Dung V, Dawson S, Arctander P, Mackinnon J (1994) Science and Technology News. Hanoi: Forest Inventory and Planning Institute, 4–13 March.
Wang YX (1998) Gongshan muntjac (Muntiacus gongshanensis). In Wang S, ed., China Red Data Book of Endangered Animals (Mammalia). Beijing: Science Press, pp. 287–288.
Wang YX (2003) A Complete Checklist of Mammal Species and Subspecies in China (A taxonomic and Geographic Reference). China Forestry Publishing House.
Wang W, Lan H (2000) Rapid and parallel chromosomal number reductions in muntjac deer inferred from mitochondrial DNA phylogeny. Mol Biol Evol 17: 1326–1333.
White MJD (1978) Modes of Speciation. San Francisco: Freeman.
Wurster DH, Benirschke K (1967) Chromosome studies in some deer, the springbok, and the pronghorn, with notes on the placentation in deer. Cytologia 32: 273–285.
Wurster DH, Atkin NB (1972) Muntjac chromosomes: a new karyotype for Muntiacus muntjak. Experimentia 28: 972–973.
Wurster DH, Benirschke K (1970) Indian muntjac, Muntiacus muntjak: a deer with a low diploid chromosome number. Science 168: 1364–1366.
Yang F (1998) Chromosome evolution of the muntjacs: inferences from molecular cytogenetics. Thesis (PhD). University of Cambridge.
Yang F, Carter NP, Shi L, Ferguson-Smith MA (1995) A comparative study of karyotypes of muntjacs by chromosome painting. Chromosoma 103: 642–652.
Yang F, Müller S, Just R, Ferguson-Smith MA, Wienberg J (1997a) Comparative chromosome painting in mammals: human and the Indian muntjac (Muntiacus muntjak vaginalis). Genomics 39: 396–401.
Yang F, O’Brien PCM, Wienberg J, Ferguson-Smith MA (1997b) A reappraisal of the tandem fusion theory of karyotype evolution in the Indian muntjac using chromosome painting. Chromosome Res 5: 109–117.
Yang F, O’Brien PCM, Wienberg J, Ferguson-Smith MA (1997c) Evolution of the black muntjac (Muntiacus crinifrons) karyotype revealed by comparative chromosome painting. Cytogenet Cell Genet 76: 159–163.
Yang F, O’Brien PCM, Wienberg J, Neitzel H, Lin CC, Ferguson-Smith MA (1997d) Chromosomal evolution of the Chinese muntjac (Muntiacus reevesi). Chromosoma 106: 37–43.
Yang F, Graphodatsky AS, O’Brien PCM et al. (2000) Reciprocal chromosome painting illuminates the history of genome evolution of the domastic cat, dog and human. Chromosome Res 8: 393–404.
Zhang CZ, Sheng HL, Lu HG (1984) On Fea’s muntjac from Xizang (Tibet), China. Acta Theriol Sinica 4: 88–106.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Huang, L., Wang, J., Nie, W. et al. Tandem chromosome fusions in karyotypic evolution of Muntiacus: evidence from M. feae and M. gongshanensis . Chromosome Res 14, 637–647 (2006). https://doi.org/10.1007/s10577-006-1073-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-006-1073-2