Abstract
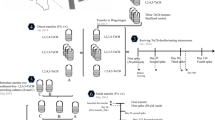
Anaerobic reductive dechlorination of hexachlorobenzene (HCB) and three isomers of tetrachlorobenzene (TeCB) (1,2,3,4-, 1,2,3,5- and 1,2,4,5-TeCB) was investigated in microcosms containing chloroaromatic contaminated river sediment. All chlorobenzenes were dechlorinated to dichlorobenzene (DCB) or monochlorobenzene. From the sediment, a methanogenic sediment-free culture was obtained which dechlorinated HCB, pentachlorobenzene, three TeCB isomers, three trichlorobenzene (TCB) isomers (1,2,3-, 1,2,4- and 1,3,5-TCB) and 1,2-DCB. Dechlorination involved multiple pathways including the removal of doubly flanked, singly flanked and isolated chlorine substituents. 454-pyrosequencing of partial bacterial 16S rRNA genes amplified from selected chlorobenzene dechlorinating sediment-free enrichment cultures revealed the presence of a variety of bacterial species, including Dehalobacter and Dehalococcoides mccartyi, that were previously documented as organohalide respiring bacteria. A genus with apparent close relationship to Desulfitobacterium that also has been associated with organohalide respiration, composed the major fraction of the operational taxonomic units (OTUs). Another major OTU was linked with Sedimentibacter sp., a genus that was previously identified in strict co-cultures of consortia reductively dehalogenating chlorinated compounds. Our data point towards the existence of multiple interactions within highly chlorinated benzene dechlorinating communities.







Similar content being viewed by others
References
Adrian L, Görisch H (2002) Microbial transformation of chlorinated benzenes under anaerobic conditions. Res Microbiol 153:131
Adrian L, Szewzyk U, Wecke J, Görisch H (2000) Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580
Alexander M (1965) Biodegradation: problems of molecular recalcitrance and microbial fallibility. Adv Appl Microbiol 7:35–80
Baena S, Fardeau ML, Labat M et al (1998) Aminobacterium colombiense gen. nov. sp. nov., an amino acid-degrading anaerobe isolated from anaerobic sludge. Anaerobe 4:241–250
Barber JL, Sweetman AJ, van Wijk D, Jones KC (2005) Hexachlorobenzene in the global environment: emissions, levels, distribution, trends and processes. Sci Total Environ 349:1–44
Bedard DL, Ritalahti KM, Löffler FE (2007) The Dehalococcoides population in sediment-free mixed cultures metabolically dechlorinates the commercial polychlorinated biphenyl mixture aroclor 1260. Appl Environ Microbiol 73:2513–2521
Beurskens JEM, Dekker CGC, van den Heuvel H et al (1994) Dechlorination of chlorinated benzenes by an anaerobic microbial consortium that selectively mediates the thermodynamic most favorable reactions. Environ Sci Technol 28:701–706
Bosma TNP, van der Meer JR, Schraa G et al (1988) Reductive dechlorination of all trichloro- and dichlorobenzene isomers. FEMS Microbiol Lett 53:223–229
Bouchard B, Beaudet R, Villemur R et al (1996) Isolation and characterization of Desulfitobacterium frappieri sp. nov., an anaerobic bacterium which reductively dechlorinates pentachlorophenol to 3-chlorophenol. Int J Syst Bacteriol 46:1010–1015
Breitenstein A, Wiegel J, Haertig C et al (2002) Reclassification of Clostridium hydroxybenzoicum as Sedimentibacter hydroxybenzoicus gen. nov., comb. nov., and description of Sedimentibacter saalensis sp. nov. Int J Syst Evol Microbiol 52:801–807
Bunge M, Wagner A, Fischer M et al (2008) Enrichment of a dioxin-dehalogenating Dehalococcoides species in two-liquid phase cultures. Environ Microbiol 10:2670–2683
Chang B-V, Chen Y-M, Yuan S-Y, Wang Y-S (1997) Reductive dechlorination of hexachlorobenzene by an anaerobic mixed culture. Water Air Soil Pollut 100:25–32
Cichocka D, Nikolausz M, Haest PJ, Nijenhuis I (2010) Tetrachloroethene conversion to ethene by a Dehalococcoides-containing enrichment culture from Bitterfeld. FEMS Microbiol Ecol 72:297–310
Cock PJA, Antao T, Chang JT et al (2009) Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423
Cravo-Laureau C, Matheron R, Joulian C et al (2004) Desulfatibacillum alkenivorans sp. nov., a novel n-alkene-degrading, sulfate-reducing bacterium, and emended description of the genus Desulfatibacillum. Int J Syst Evol Microbiol 54:1639–1642
Dijk JA, Breugelmans P, Philips J et al (2008) Catalyzed reporter deposition-fluorescent in situ hybridization (CARD-FISH) detection of Dehalococcoides. J Microbiol Methods 73:142–147
Ding C, He J (2012) Molecular techniques in the biotechnological fight against halogenated compounds in anoxic environments. Microbiol Biotechnol 5:347–367
Dipippo JL, Nesbø CL, Dahle H et al (2009) Kosmotoga olearia gen. nov., sp. nov., a thermophilic, anaerobic heterotroph isolated from an oil production fluid. Int J Syst Evol Microbiol 59:2991–3000
Dolfing J, Harrison BK (1993) Redox and reduction potentials as parameters to predict the degradation pathway of chlorinated benzenes in anaerobic environments. FEMS Microbiol Ecol 13:23–29
Duan TH, Adrian L (2013) Enrichment of hexachlorobenzene and 1,3,5-trichlorobenzene transforming bacteria from sediments in Germany and Vietnam. Biodegradation 24:513–520
Duhamel M, Edwards EA (2007) Growth and Yyelds of dechlorinators, acetogens, and methanogens during reductive dechlorination of chlorinated ethenes and dihaloelimination of 1,2-dichloroethane. Environ Sci Technol 41:2303–2310
Edgar RC, Haas BJ, Clemente JC et al (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Fathepure BZ, Tiedje JM, Boyd SA (1988) Reductive dechlorination of hexachlorobenzene to tri- and dichlorobenzenes in anaerobic sewage sludge. Appl Environ Microbiol 54:327–330
Fennell DE, Nijenhuis I, Wilson SF et al (2004) Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ Sci Technol 38:2018–2075
Field J, Sierra-Alvarez R (2008) Microbial degradation of chlorinated benzenes. Biodegradation 19:463–480
Finneran KT (2002) Desulfitobacterium metallireducens sp. nov., an anaerobic bacterium that couples growth to the reduction of metals and humic acids as well as chlorinated compounds. Int J Syst Evol Microbiol 52:1929–1935
Fung JM, Weisenstein BP, Mack EE et al (2009) Reductive dehalogenation of dichlorobenzenes and monochlorobenzene to benzene in microcosms. Environ Sci Technol 43:2302–2307
Gerritse J, Renard V, Pedro Gomes TM et al (1996) Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols. Arch Microbiol 165:132–140
Gunsalus RP, Romesser JA, Wolfe RS (1978) Preparation of coenzyme M analogs and their activity in the methyl coenzyme M reductase system of Methanobacterium thermoautotrophicum. Biochemistry 17:2374–2377
He J, Sung Y, Krajmalnik-Brown R et al (2005) Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ Microbiol 7:1442–1450
Heimann AC, Batstone DJ, Jakobsen R (2006) Methanosarcina spp. drive vinyl chloride dechlorination via interspecies hydrogen transfer. Appl Environ Microbiol 72:2942–2949
Holliger C, Schraa G, Stams AJ, Zehnder AJ (1992) Enrichment and properties of an anaerobic mixed culture reductively dechlorinating 1,2,3-trichlorobenzene to 1,3-dichlorobenzene. Appl Environ Microbiol 58:1636–1644
Holliger C, Schraa G, Stams AJM, Zehnder AJB (1993) A highly purified enrichment culture couples the reductive dechlorination of tetrachloroethene to growth. Appl Environ Microbiol 59:2991–2997
Hölscher T, Lisec J, Baani M et al (2010) Bacterial cultures preferentially removing singly flanked chlorine substituents from chlorobenzenes. Environ Sci Technol 44:8936–8942
Hug LA, Maphosa F, Leys D et al (2013) Overview of organohalide-respiring bacteria and a proposal for a classification system for reductive dehalogenases. Philos Trans R Soc B 368:20120322
Imachi H, Sakai S, Ohashi A et al (2007) Pelotomaculum propionicicum sp. nov., an anaerobic, mesophilic, obligately syntrophic, propionate-oxidizing bacterium. Int J Syst Evol Microbiol 57:1487–1492
Jayachandran G, Görisch H, Adrian L (2003) Dehalorespiration with hexachlorobenzene and pentachlorobenzene by Dehalococcoides sp. strain CBDB1. Arch Microbiol 180:411–416
Kaufhold T, Schmidt M, Cichocka D et al (2013) Dehalogenation of diverse halogenated substrates by a highly enriched Dehalococcoides-containing culture derived from the contaminated mega-site in Bitterfeld. FEMS Microbiol Ecol 83:176–188
Kim O-S, Cho Y-J, Lee K et al (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Lee Y-J, Romanek CS, Mills GL et al (2006) Gracilibacter thermotolerans gen. nov., sp. nov., an anaerobic, thermotolerant bacterium from a constructed wetland receiving acid sulfate water. Int J Syst Evol Microbiol 56:2089–2093
Lee J, Lee TK, Löffler FE, Park J (2011) Characterization of microbial community structure and population dynamics of tetrachloroethene-dechlorinating tidal mudflat communities. Biodegradation 22:687–698
Li Z, Inoue Y, Suzuki D et al (2013) Long-term anaerobic mineralization of pentachlorophenol in a continuous-flow system using only lactate as an external nutrient. Environ Sci Technol 47:1534–1541
Löffler FE, Yan J, Ritalahti KM et al (2013) Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and famil. Int J Syst Evol Microbiol 63:625–635
Maphosa F, de Vos WM, Smidt H (2010) Exploiting the ecogenomics toolbox for environmental diagnostics of organohalide-respiring bacteria. Trends Biotechnol 28:308–316
Maphosa F, van Passel MWJ, de Vos WM, Smidt H (2012) Metagenome analysis reveals yet unexplored reductive dechlorinating potential of Dehalobacter sp. E1 growing in co-culture with Sedimentibacter sp. Environ Microbiol Rep 4:604–616
Masunaga S, Susarla S, Yonezawa Y (1996) Dechlorination of chlorobenzenes in anaerobic estuarine sediment. Water Sci Technol 33:173–180
May HD, Miller GS, Kjellerup BV, Sowers KR (2008) Dehalorespiration with polychlorinated biphenyls by an anaerobic ultramicrobacterium. Appl Environ Microbiol 74:2089–2094
McInerney MJ, Bryant MP, Hespell RB, Costerton JW (1981) Syntrophomonas wolfei gen. nov. sp. nov., an anaerobic, syntrophic, fatty acid-oxidizing bacterium. Appl Environ Microbiol 41:1029–1039
Middeldorp PJM, De Wolf J, Zehnder AJB, Schraa G (1997) Enrichment and properties of a 1,2,4-trichlorobenzene-dechlorinating methanogenic microbial consortium. Appl Environ Microbiol 63:1225–1229
Muyzer G, De Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nelson JL, Fung JM, Cadillo-Quiroz H et al (2011) A role for Dehalobacter spp. in the reductive dehalogenation of dichlorobenzenes and monochlorobenzene. Environ Sci Technol 45:6806–6813
Nijenhuis I, Stelzer N, Kästner M, Richnow H-H (2007) Sensitive detection of anaerobic monochlorobenzene degradation using stable isotope tracers. Environ Sci Technol 41:3836–3842
Nowak J, Kirsch NH, Hegemann W, Stan HJ (1996) Total reductive dechlorination of chlorobenzenes to benzene by a methanogenic mixed culture enriched from Saale river sediment. Appl Microbiol Biotechnol 45:700–709
Pöritz M, Goris T, Wubet T et al (2013) Genome sequences of two dehalogenation specialists—Dehalococcoides mccartyi strains BTF08 and DCMB5 enriched from the highly polluted Bitterfeld region. FEMS Microbiol Lett 343:101–104
Quistorff AS (1999) Microbially mediated reductive dechlorination of dichlorobenzene. Dissertation, Cornell University, Ithaca
Ramanand K, Balba MT, Duffy J (1993) Reductive dehalogenation of chlorinated benzenes and toluenes under methanogenic conditions. Appl Environ Microbiol 59:3266–3272
Rosenzweig N, Tiedje JM, Quensen JF et al (2012) Microbial communities associated with potato common scab-suppressive soil determined by pyrosequencing analyses. Plant Dis 96:718–725
Schaefer CE, Condee CW, Vainberg S, Steffan RJ (2009) Bioaugmentation for chlorinated ethenes using Dehalococcoides sp.: comparison between batch and column experiments. Chemosphere 75:141–148
Schloss PD, Westcott SL, Ryabin T et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Seshadri R, Adrian L, Fouts DE et al (2005) Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105–108
Smits THM, Devenoges C, Szynalski K et al (2004) Development of a real-time PCR method for quantification of the three genera Dehalobacter, Dehalococcoides, and Desulfitobacterium in microbial communities. J Microbiol Methods 57:369–378
Sokolova TG, Kostrikina NA, Chernyh NA et al (2002) Carboxydocella thermautotrophica gen. nov., sp. nov., a novel anaerobic, CO-utilizing thermophile from a Kamchatkan hot spring. Int J Syst Evol Microbiol 52:1961–1967
Taş N, van Eekert MHA, Schraa G et al (2009) Tracking functional guilds: “Dehalococcoides” spp. in European river basins contaminated with hexachlorobenzene. Appl Environ Microbiol 75:4696–4704
Taş N, van Eekert MHA, de Vos WM, Smidt H (2010) The little bacteria that can—diversity, genomics and ecophysiology of “Dehalococcoides” spp. in contaminated environments. Microbiol Biotechnol 3:389–402
Van de Pas B, Harmsen H, Raangs G et al (2001) A Desulfitobacterium strain isolated from human feces that does not dechlorinate chloroethenes or chlorophenols. Arch Microbiol 175:389–394
Van Eekert MHA, Stams AJM, Field JA, Schraa G (1999) Gratuitous dechlorination of chloroethanes by methanogenic granular sludge. Appl Microbiol Biotechnol 51:46–52
Villemur R (2013) The pentachlorophenol-dehalogenating Desulfitobacterium hafniense strain PCP-1. Philos Trans R Soc B 368:20120319
Wu Q, Milliken CE, Meier GP et al (2002) Dechlorination of chlorobenzenes by a culture containing bacterium DF-1, a PCB dechlorinating microorganism. Environ Sci Technol 36:3290–3294
Yu S, Semprini L (2004) Kinetics and modeling of reductive dechlorination at high PCE and TCE concentrations. Biotechnol Bioeng 88:451–464
Zellner G, Stackebrandt E, Messner P et al (1989) Methanocorpusculaceae fam. nov., represented by Methanocorpusculum parvum, Methanocorpusculum sinense spec. nov. and Methanocorpusculum bavaricum spec. nov. Arch Microbiol 151:381–390
Zhang H, Ziv-El M, Rittmann BE, Krajmalnik-Brown R (2010) Effect of dechlorination and sulfate reduction on the microbial community structure in denitrifying membrane-biofilm reactors. Environ Sci Technol 44:5159–5164
Ziv-El M, Popat SC, Cai K et al (2012) Managing methanogens and homoacetogens to promote reductive dechlorination of trichloroethene with direct delivery of H2 in a membrane biofilm reactor. Biotechnol Bioeng 109:2200–2210
Acknowledgments
We thank the Flemish Environmental and Technology Innovation Platform for funding this research as part of the MIP project. Furthermore, we thank P.J. Haest for his help with the real-time qPCR and S. Ruyters for assistance with analyzing 454-pyrosequencing data.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
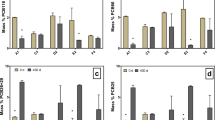
Concentration of CBs in killed controls of Kanaal Ieper-Ijzer sediment microcosms. Microcosms were amended with a HCB, b 1,2,3,4-TeCB, c 1,2,3,5-TeCB, and d 1,2,4,5-TeCB. Symbols indicate: HCB (X), 1,2,3,4-TeCB (filled triangle), 1,2,3,5-TeCB (open square) and 1,2,4,5-TeCB (filled triangle). Presented data points are averages of two replicate microcosms. Standard error bars are indicated. Supplementary material 1 (PPT 197 kb)
Supplementary Table 1
Selected representative OTU sequences and their closest match in GenBank. The OTUs presented are the first 26 in number and represent 93 % of the sequences of the dataset. The dataset consisted of 7,376 sequences, organised into 161 OTUs (defined by a 3 % sequence dissimilarity cut-off). The coverage is the percent of the query sequence that overlaps the subject sequence in GenBank and the similarity is the percent similarity between the query and subject sequences over the length of the coverage area. Supplementary material 2 (DOC 67 kb)
Rights and permissions
About this article
Cite this article
Vandermeeren, P., Herrmann, S., Cichocka, D. et al. Diversity of dechlorination pathways and organohalide respiring bacteria in chlorobenzene dechlorinating enrichment cultures originating from river sludge. Biodegradation 25, 757–776 (2014). https://doi.org/10.1007/s10532-014-9697-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-014-9697-y