Abstract
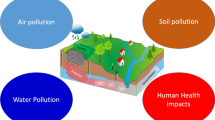
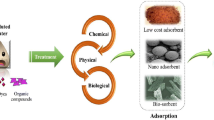
Intensive industrialisation, inadequate disposal, large-scale manufacturing activities and leaks of organic compounds have resulted in long-term persistent sources of contamination of soil and groundwater. This is a major environmental, policy and health issue because of adverse effects of contaminants on humans and ecosystems. Current technologies for remediation of contaminated sites include chemical and physical remediation, incineration and bioremediation. With recent advancements, bioremediation offers an environmentally friendly, economically viable and socially acceptable option to remove contaminants from the environment. Three main approaches of bioremediation include use of microbes, plants and enzymatic remediation. All three approaches have been used with some success but are limited by various confounding factors. In this paper, we provide a brief overview on the approaches, their limitations and highlights emerging technologies that have potential to revolutionise the enzymatic and plant-based bioremediation approaches.


Similar content being viewed by others
References
Abhilash PC, Singh HB, Powell JR, Singh BK (2012) Plant–microbe interactions: novel applications for exploitation in multi-purpose remediation technologies. Trend Biotechnol 30:416–420
Beil S, Mason JR, Timmis KN, Pieper DH (1998) Identification of chlorobenzene dioxygenase sequence elements involved in dechlorination of 1,2,4,5-tetrachlorobenzene. J Bacteriol 180:5520–5528
Black H (1995) Absorbing possibilities: phytoremediation. Environ Health Perspect 103:1106
Blasco R, Wittich RM, Megharaj M, Timmis KN, Pieper DH (1995) From xenobiotic to antibiotic: formation of protoanemonin from 4-chlorocatechol by enzymes of the 3-oxoadipate pathway. J Biol Chem 270:29229–29235
Brar SK, Verma M, Surampalli RY, Misra K, Tyagi RD, Meunier N, Blais JF (2006) Bioremediation of hazardous wastes—a review. Pract Period Hazard Toxic Radioact Waste Manag 10:59
Chaudhry T, Hayes W, Khan A, Khoo C (1998) Phytoremediation—focusing on accumulator plants that remediate metal-contaminated soils. Australas J Ecotoxicol 4:37–51
Cluis C (2004) Junk-greedy greens: phytoremediation as a new option for soil decontamination. BioTeach J 2:61–67
Curtis TP, Sloan WT, Scannell JW (2002) Estimating prokaryotic diversity and its limits. Proc Nat Acad Sci USA 99:10494–10497
Erb RW, Eichner CA, Wagner-Döbler I, Timmis KN (1997) Bioprotection of microbial communities from toxic phenol mixtures by a genetically designed pseudomonad. Nat Biotechnol 15:378–382
Fan X, Liu X, Huang R, Liu Y (2012) Identification and characterization of a novel thermostable pyrethroid-hydrolyzing enzyme isolated through metagenomic approach. Microb Cell Fact 11:33–37
Gerhardt KE, Huang X-D, Glick BR, Greenberg BM (2009) Phytoremediation and rhizoremediation of organic soil contaminants: potential and challenges. Plant Sci 176:20–30
Ginolhac A, Jarrin C, Gillet B, Robe P, Pujic P, Tuphile K, Bertrand H, Vogel TM, Perrière G, Simonet P, Nalin R (2004) Phylogenetic analysis of polyketide synthase I domains from soil metagenomic libraries allows selection of promising clones. Appl Environ Microbiol 70:5522–5527
Gulmaraes BCM, Arends JBA, Van der Ha D, Boon N, Verstraetes W (2010) Microbial service and their management: recent progress in soil bioremediation technologies. Appl Soil Ecol 46:157–167
Guo HH, Choe J, Loeb LA (2004) Protein tolerance to random amino acid change. Proc Natl Acad Sci USA 101:9205
Hadacek F (2002) Secondary metabolites as plant traits: current assessment and future perspectives. Crit Rev Plant Sci 21:273–322
Hartman Jr WJ (1975) An evaluation of land treatment of municipal wastewater and physical siting of facility installations. DTIC Document
Hatzinger PB, Whittier MC, Arkins MD, Bryan CW, Guarini WJ (2002) In situ and ex-situ bioremediation options for treating perchlorate in groundwater. Remediat J 12:69–86
Hazen TC, Dubinsky EA, DeSantis TZ et al (2010) Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208
Holloway P, Knoke KL, Trevors JT, Lee H (1998) Alteration of the substrate range of haloalkane dehalogenase by site directed mutagenesis. Biotechnol Bioeng 59:520–523
Jones KC, de Voogt P (1999) Persistent organic pollutants (POPs): state of the science. Environ Pollut 100:209–222
Ju KS, Parales RE (2006) Control of substrate specificity by active-site residues in nitrobenzene dioxygenase. Appl Environ Microbiol 72:1817–1824
Juwarkar A, Singh S, Mudhoo A (2010) A comprehensive overview of elements in bioremediation. Rev Environ Sci Biotechnol 9:215–288
Karns JS, Hapeman CJ, Mulbry WW, Ahrens EH, Shelton DR (1998) Biotechnology for the elimination of agrochemical wastes. HortScience 33:626–631
Kind S, Jeong WK, Schröder H, Zelder O, Wittmann C (2010) Identification and elimination of the competing N-acetyldiaminopentane pathway for improved production of diaminopentane by Corynebacterium glutamicum. Appl Environ Microbiol 76:5175–5180
Koffas M, Roberge C, Lee K, Stephanopoulos G (1999) Metabolic engineering. Annu Rev Biomed Eng 1:535–557
Kuchner O, Arnold FH (1997) Directed evolution of enzyme catalysts. Trend Biotechnol 15:523–530
Kumar PBAN, Dushenkov V, Motto H, Raskin I (1995) Phytoextraction: the use of plants to remove heavy metals from soils. Environ Sci Technol 29:1232–1238
Kurumata M, Takahashi M, Sakamotoa A, Ramos JL, Nepovim A, Vanek T, Hirata T, Morikawa H (2005) Tolerance to, and uptake and degradation of 2,4,6-trinitrotoluene (TNT) are enhanced by the expression of a bacterial nitroreductase gene in Arabidopsis thaliana. Z Naturforsch 60:272–278
Lee JY, Jung KH, Choi SH, Kim HS (1995) Combination of the tod and the tol pathways in redesigning a metabolic route of Pseudomonas putida for the mineralization of a benzene, toluene, and p-xylene mixture. Appl Environ Microbiol 61:2211–2217
Lee JH, Hwang ET, Kim BC, Lee SM, Sang BI, Choi YS, Kim J, Gu MB (2007) Stable and continuous long-term enzymatic reaction using an enzyme—nanofiber composite. Appl Microbiol Biotechnol 75:1301–1307
Li T, Guo S, Wu B, Li F, Niu Z (2010) Effect of electric intensity on the microbial degradation of petroleum pollutants in soil. J Environ Sci 22:1381–1386
Marconi AM, Kieboom J, De Bont JAM (1997) Improving the catabolic functions in the toluene-resistant strain Pseudomonas putida s12. Biotechnol Lett 19:603–606
McCutcheon SC, Schnoor JL (2003) Phytoremediation: transformation and control of contaminants, vol 118. LibreDigital
McMahon V, Garg A, Aldred D, Hobbs G, Smith R, Tothill I (2008) Composting and bioremediation process evaluation of wood waste materials generated from the construction and demolition industry. Chemosphere 71:1617–1628
Megharaj M, Ramakrishnan B, Venkateswarlu K, Sethunathan N, Naidu R (2011) Bioremediation approaches for organic pollutants: a critical perspective. Environ Int 37:1362–1375
Mulbry W, Ahrens E, Karns J (1998) Use of a field scale biofilter for the degradation of the organophosphate insecticide coumaphos in cattle dip wastes. Pestic Sci 52:268–274
Paul D, Pandey G, Pandey J, Jain RK (2005) Accessing microbial diversity for bioremediation and environmental restoration. Trend Biotechnol 23:135–142
Pazirandeh M, Wells BM, Ryan RL (1998) Development of bacterium-based heavy metal biosorbents: enhanced uptake of cadmium and mercury by Escherichia coli expressing a metal binding motif. Appl Environ Microbiol 64:4068–4072
Pei X-H, Zhan X-H, Wang S-M, Lin Y-S, Zhou L-X (2010) Effects of a biosurfactant and a synthetic surfactant on phenanthrene degradation by a Sphingomonas strain. Pedosphere 20:771–779
Pulford I, Watson C (2003) Phytoremediation of heavy metal-contaminated land by trees—a review. Environ Int 29:529–540
Ramos JL, Gonzalez-Perez MM, Caballero A, van Dillewijn P (2005) Bioremediation of polynitrated aromatic compounds: plants and microbes put up a fight. Curr Opin Biotechnol 16:275–281
Rascio N, Navari-Izzo F (2011) Heavy metal hyperaccumulating plants: how and why do they do it? And what makes them so interesting? Plant Sci 180:169–181
Renukaradhya M, Shah AI et al (2010) Isolation of a novel gene encoding a 3,5,6-trichloro-2-pyridinol degrading enzyme from a cow rumen metagenomic library. Biodegradation 21:565–573
Rojo F, Pieper DH, Engesser KH, Knackmuss HJ, Timmis KN (1987) Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science 238:1395–1398
Rylott EL, Jackson RG, Edwards J, Womack GL, Seth-Smith HMB, Rathbone DA, Strand SE, Bruce NC (2006) An explosive-degrading cytochrome P450 activity and its targeted application for the phytoremediation of RDX. Nat Biotechnol 24:216–219
Salt DE, Blaylock M, Kumar NPBA, Dushenkov V, Ensley BD, Chet I, Raskin I (1995) Phytoremediation: a novel strategy for the removal of toxic metals from the environment using plants. Nat Biotechnol 13:468–474
Schnoor JL, Light LA, McCutcheon SC, Wolfe NL, Carreia LH (1995) Phytoremediation of organic and nutrient contaminants. Environ Sci Technol 29:318–323
Scott C, Begley C, Taylor MJ, Pandey G, Momiroski V, French N, Brearley B, Kotsonis SE, Selleck MJ, Carino FA, Bajet CM, Clarke C, Oakeshott JG, Russell RJ (2011) Free enzyme bioremediation of pesticides. ACS symposium series book 1075:155–174
Shao Z, Zhao H, Giver L, Arnold FH (1998) Random-priming in vitro recombination: an effective tool for directed evolution. Nucleic Acids Res 26:681–683
Singer AC, Crowley DE, Thompson IP (2003) Secondary plant metabolites in phytoremediation and biotransformation. Trend Biotechnol 21:123–130
Singer AC, Thompson IP, Bailey MJ (2004) The tritrophic trinity: a source of pollutant-degrading enzymes and its implications for phytoremediation. Curr Opin Microbiol 7:239–244
Singh BK (2009) Organophosphorus-degrading bacteria: ecology and industrial applications. Nat Rev Microbiol 7:156–164
Singh BK (2010) Exploring microbial diversity for biotechnology: the way forward. Trend Biotechnol 28:111–116
Singh BK, Walker A (2006) Microbial degradation of organophosphorus compounds. FEMS Microbiol Rev 30:428–471
Singh BK, Campbell C, Sorensen SJ, Zhou J (2009) Soil genomics is the way forward. Nat Rev Microbiol 7:756
Strong LC, McTavish H, Sadowsky MJ, Wackett LP (2000) Field-scale remediation of atrazine-contaminated soil using recombinant Escherichia coli expressing atrazine chlorohydrolase. Environ Microbiol 2:91–98
Sul WJ, Park J, Quensen JF III, Rodrigues JLM, Seliger L, Tsoi TV, Zylstra GJ, Tiedje JM (2009) DNA-stable isotope probing integrated with metagenomics for retrieval of biphenyl dioxygenase genes from polychlorinated biphenyl-contaminated river sediment. Appl Environ Microbiol 75:5501–5506
Tu C, Ma LQ, Zhang W, Cai Y, Harris WG (2003) Arsenic species and leachability in the fronds of the hyperaccumulator Chinese brake (Pteris vittata L.). Environ Pollut 124:223–230
Velkov VV (2001) Stress-induced evolution and the biosafety of genetically modified microorganisms released into the environment. J Biosci 26:667–683
Walker AW, Keasling JD (2002) Metabolic engineering of Pseudomonas putida for the utilization of parathion as a carbon and energy source. Biotechnol Bioeng 78:715–721
Wang XX et al (2008) Phytodegradation of organophosphorus compounds by transgenic plants expressing a bacterial organophosphorus hydrolase. Biochem Biophys Res Commun 365:453–458
Watanabe K (2001) Microorganisms relevant to bioremediation. Curr Opin Biotechnol 12:237–241
Wei S, Zhou QX (2006) Phytoremediation of cadmium-contaminated soils by Rorippa globosa using two-phase planting. Environ Sci Pollut Res 13:151–155
Yergeau E, Arbour M, Brousseau R, Juck D, Lawrence JR, Masson L, Whyte LG, Greer CW (2009) Microarray and real-time PCR analyses of the responses of high-arctic soil bacteria to hydrocarbon pollution and bioremediation treatments. Appl Environ Microbiol 75:6258–6267
Zhao H, Giver L, Shao Z, Affholter JA, Arnold FH (1998) Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat Biotechnol 16:258–261
Zhao F, Lombi E, Breedon T (2000) Zinc hyperaccumulation and cellular distribution in Arabidopsis halleri. Plant Cell Environ 23:507–514
Acknowledgments
This paper is based on plenary presentation by BKS at Cleanup 2011 conference, Adelaide, Australia. BKS acknowledge funding from Hawkesbusry Institute for Environment, Grains Research Development Corporation and Cotton Research Development Corporation. DGK acknowledges funding of his group by the Postgraduate Program “Biotechnology-Quality assessment in Nutrition and the Environment”, Department of Biochemistry and Biotechnology, University of Thessaly.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rayu, S., Karpouzas, D.G. & Singh, B.K. Emerging technologies in bioremediation: constraints and opportunities. Biodegradation 23, 917–926 (2012). https://doi.org/10.1007/s10532-012-9576-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-012-9576-3