Abstract
Identifying sources of range expansions after an introduction event and understanding the species dispersal are essential for effective management of invasive species. In a unique study system we investigated the spread and distribution of genetic diversity subsequent to a known colonization event and in the light of the well-known biology of the rapidly expanding Roesel’s bush-cricket (Metrioptera roeselii), a species that is non-native in Sweden. Using eight microsatellite markers we analyzed genetic variation in 837 individuals collected at 29 sites across the species total range in central Sweden to verify the species local origin and to determine how the species known dispersal biology affect the population genetic structure throughout its range. We found that unique allelic richness was highest in a site approximately 16 km southwest of the previously suggested site of establishment, pointing towards a site of introduction close to a royal stud farm from where it is known that animals have been imported from Europe. Despite the species rapid expansion, genetic diversity in the core of the distribution was higher than in the populations at the range margin. Bayesian cluster analyses also revealed that genetic structuring was more pronounced in the range margin, indicating the occurrence of dichotomous dispersal behaviour of the species with occasions of rare long distance events. Our study shows that good sampling design and appropriate choice of genetic markers can help to identify species local origin and explain genetic patterns that arise during range expansions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alteration of habitats and increased international trade are factors that contribute to the colonisation success of exotic species with potentially cascading effects on native species persistence, composition of local communities and ecosystem functioning (Sax et al. 2005; Suarez and Tsutsui 2008; Keller and Taylor 2010). Early intervention is crucial to successfully manage the expansion of non-native species in a region and this is aided by knowledge of the species origin, its ecology and its dispersal behaviour (Hulme 2006). Unless deliberate and post-introduction censuses are done, species colonisations and spread are initially difficult to detect because they usually consist of rare events and/or occur over large spatial and temporal scales (Puth and Post 2005). As a result, range expansions remain usually unnoticed until the species has become locally abundant (Sakai et al. 2001). Identifying the source of introduction, dispersal routes and the patterns of spread are essential for understanding the dynamics of species range expansions and the development of effective management strategies (Puth and Post 2005; Wilson et al. 2009; Ronnås et al. 2011).
However, studying the distribution change of non-native insects over large spatial and temporal scales has its challenges due to the insects small body size, rapid reproduction and often complex dispersal behaviour. The use of neutral molecular markers and population genetic inferences has become a popular approach to investigate the history of populations and quantifying dispersal patterns (Brown 2004; Lawson Handley et al. 2011). Spatial and temporal variation of genetic structure arises as new populations are founded during range expansion events (Ibrahim et al. 1996) and are shaped by migration of individuals between populations. The amount of genetic variation in newly founded populations depends on the number and origin of founding individuals and subsequent immigrants (Le Corre and Kremer 1998; Dlugosch and Parker 2008; Kaňuch et al. 2014). Further, subsequent changes in genetic diversity within and among populations are influenced by the combined effects of the species dispersal behaviour and environmental factors. If a species spreads gradually and in a stepping-stone model fashion (Wright 1943; Kimura and Weiss 1964), one expects to find a correlation between the geographical distance separating populations and the genetic distance estimated from genetic markers. This is referred to as an isolation-by-distance pattern (Wright 1943). A more complex dispersal behaviour of a species is expected to result in a weak isolation-by-distance pattern and a spatially more complex genetic structure. One example of complex dispersal is the combination of long distance dispersal events by single colonizers that create genetic structures with populations having low and altered diversity due to local founder effects (Nichols and Hewitt 1994; Bialozyt et al. 2006) combined with short distance and frequent dispersal, as shown in dimorphic insects (Roff 1986).
There is a need to couple specific biological traits with historical events in order to fully understand population genetic patterns of expanding species where the biological knowledge is less well known (Kirk et al. 2013). In order to add to the knowledge of how introductions to a new area and subsequent expansion we used a unique study system including a population of an introduced and rapidly expanding species (Metrioptera roeselii) where the history is well known (de Jong and Kindvall 1991; Preuss et al. 2011; Kaňuch et al. 2013) and where the species biology and dispersal behaviour has been extensively studied (Berggren et al. 2001; Berggren 2004; Simmons and Thomas 2004; Hochkirch and Damerau 2009; Holzhauer et al. 2009; Lange et al. 2010). This combination allows us to investigate fundamental population genetic processes and a non-native species dispersal patterns during range expansion (Kirk et al. 2013).
Metrioptera roeselii was first sighted in Sweden in 1882 near the city of Västerås at the shore of Lake Mälaren. The species was assumed of having been introduced via sea cargo (de Jong and Kindvall 1991) which is supported by findings that trace the species origin to northern Germany and/or Poland (Kanuch et al. 2013). In Sweden M. roeselii went largely unnoticed for nearly 100 years and local spread was first noted in 1970 (Idar 1973). Comprehensive surveys were conducted during 1989–1990 and 2008–2010 resulting in detailed documentation of range expansion in the region (de Jong and Kindvall 1991; Preuss et al. 2011). Over the last 20 years the species has continued to spread rapidly at an estimated rate of 1–3 km/year (Preuss et al. 2014) resulting in an observed shift of the range margin by more than 50 km (Preuss et al. 2011). By 2010, the species distribution in the Mälaren region (59°35′N, 16°30′E) covered an area of approximately 120 × 140 km (Fig. 1). Metrioptera roeselii is a wing dimorphic species with dichotomous dispersal behaviour (Simmons and Thomas 2004; Poniatowski and Fartmann 2011). Most individuals (usually ca. 98 %) are short-winged and disperse short distances by walking and jumping (Vickery 1965; Wissmann et al. 2009). However, favourable weather conditions (mild springs and hot summers) and high population densities may trigger the development of long-winged individuals that can colonize distant habitat patches (Simmons and Thomas 2004; Gardiner 2009). The potential impact of introduced M. roeselii on the native insect communities or ecosystem functions is yet largely unknown although there are indications of competitive dominance (cf. Berggren and Low 2004). Despite the limited evidence of negative impact on the native biota, this population constitute a useful system in which basic processes of human mediated colonisations and invasions can be studied (Allendorf and Lundquist 2003; Dlugosch and Parker 2008).
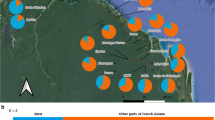
Geographic location and genetic structure of 29 Metrioptera roeselii populations sampled in the Mälaren region (59°35′N, 16°30′E), Sweden. The site number corresponds to the population number (Pop) in Table 1. Västerås (Pop 29) was previous to this study the assumed site of introduction and Ullvi (Pop 8) is the inferred site of introduction. The color of the squares depicts the ten genetic clusters identified by BAPS. The thickness of the squares outline illustrates the two genetic clusters identified by STRUCTURE
In this study we used neutral molecular genetic markers to identify M. roeselii first site of establishment and source of range expansion in central Sweden, and to investigate if the observed dispersal behaviour is mirrored in the genetic population structure as expected from theoretical modelling (Ibrahim et al. 1996; Bialozyt et al. 2006). We analysed the genetic variation and differentiation among 837 individuals from 29 locations sampled across the species total range in central Sweden (Fig. 1). We used eight polymorphic microsatellite markers to address the following three hypotheses: (1) M. roeselii was originally introduced to central Sweden at Västerås harbour and levels of genetic diversity are highest at this location. (2) Newly founded populations at the periphery of the range show lower levels of genetic diversity due to founder effects from long-distance dispersal events by long-winged individuals. (3) Subsequent and frequent short-distance dispersal by short-winged individuals reduces the genetic structure among populations situated in more central parts of the range.
Methods
Study species
Metrioptera roeselii (Hagenbach, 1822) (Orthoptera: Tettigoniidae) is a small (12–18 mm) bush-cricket commonly found in grasslands of central and northern Europe (Bellmann 2006). The species is expanding its range in several European countries (Netherlands: Kleukers et al. 1997; England: Thomas et al. 2001; Germany: Hochkirch and Damerau 2009; Wissmann et al. 2009). In northern Europe M. roeselii is both uni- and semivoltine, reproducing between July and September and overwintering as eggs in hollow grass stems and plant material (Marshall and Haes 1988). Metrioptera roeselii is an omnivorous generalist that prefers tall grassland habitats. In the agricultural landscape it colonizes extensively grazed pastures, leys, grassy field margins, ditches, and road verges (Marshall and Haes 1988; Berggren et al. 2001). Forests, crop fields and intensively grazed pastures are considered to be unsuitable habitat and urban areas are usually avoided by the species (de Jong and Kindvall 1991; Ingrisch, Köhler 1998; Wissmann et al. 2009). Short-winged individuals move on average 7.8–9 m (mean daily movement distance) and maximum 172 m/day (Kindvall et al. 1998). The measured mean activity radius of long-winged individuals is 144.4 m (Poniatowski and Fartmann 2011). High population density and favorable weather conditions can trigger the development of long-winged morphs that are capable of flight dispersal of up to 19 km and an average displacement of individuals of 10.8 km (Hochkirch and Damerau 2009).
Sampling scheme
During July and August 2010, we collected 27–30 short-winged individuals of M. roeselii per site from 29 locations across the species distribution around Lake Mälaren. The locations were situated from the core to the margin of the species distribution. The average minimum straight line (Euclidean) distance between sampling sites was 20 km and the most marginal populations were situated around 85 km from the assumed introduction site near the city of Västerås (Table 1; Fig. 1).
DNA extraction and genotyping
We isolated DNA from the femur muscle of 837 individuals using the Chelex 100 (Bio-Rad Laboratories, Inc.) extraction method (Walsh et al. 1991). We used 13 microsatellite markers that have been specifically developed for M. roeselii: Metroe05, 07, 09, 08, 19, 20, 24, 27 (Kaňuch et al. 2010) and MR2-16, 2-42, 3-12, 3-24, 3-34 (Holzhauer and Wolff 2005). The microsatellite primer pairs were arranged in two multiplex polymerase chain reaction (PCR) set-ups (Supplementary Table 1). Loci were amplified in 10 µl PCR reactions containing 1 μl DNA (~20 ng/μl), 0.3 μM of primer mixture (forward and reverse) of each locus and 2×KAPA2G Fast HotStart ReadyMix (Kapabiosystems) under the following conditions: 95 °C for 1 min, 35 cycles at 95 °C for 10 s, 56 °C for 10 s, 72 °C for 10 s and final extension at 72 °C for 1 min. Forward primers were 5′-end labelled with different fluorescent dyes (Supplementary Table 1), enabling unambiguous detection of similar sized PCR-products from co-amplified loci when analyzed on an ABI3730XL automated capillary sequencer using Gensize LIZ500 (Applied Biosystems) as a size marker. The alleles were scored using GENEMAPPER version 4.1 (Applied Biosystems). Loci with unusual peak patterns and individuals with rare alleles were analyzed twice to minimise scoring errors.
Data analyses
All loci were checked for the presence of stuttering, large allele drop out and null alleles using MICRO-CHECKER 2.2.3 (Van Oosterhout et al. 2006). We tested for linkage disequilibrium (LD) and deviation from Hardy–Weinberg equilibrium (HWE) at each locus (exact probability test with 1 000 iterations) and across all loci (Markov chain method) using GENEPOP 4.0.1 (Raymond and Rousset 1995), adjusting the significant threshold values for multiple tests using a sequential Bonferroni correction (Rice 1989). We excluded five loci (Metroe05, 07, 09, 20, MR3-12) from the subsequent analyses due to high frequency of null alleles and unclear banding patterns. Further analyses were based on eight loci (Metroe08, 16, 19, 24, 27, MR2-42, 3-24 and 3-34). For those of the remaining loci that showed evidence of null alleles in some populations, we calculated adjusted allele frequencies using the Brookfield1 correction method (Brookfield 1996) implemented in MICRO-CHECKER 2.2.3 (Van Oosterhout et al. 2006). Genetic diversity per population was described by calculating unbiased gene diversity (UHe) and inbreeding coefficient (FIS) corrected for sample size in FSTAT 2.9.3 (Goudet 2001). Mean allelic richness (AR) and mean private allelic richness (PAR) standardized for sample size (N = 26) were calculated using ADZE (Szpiech et al. 2008). Differences in levels of genetic variation between populations found within the core range versus the marginal were tested with a Mann–Whitney U test applied on the values averaged across loci. Populations found within a radius of 70 km were defined as core populations and those found further away (70–100 km) as range margin populations. To test for increasing variability in measures of genetic diversity and inbreeding from the centre of the range towards the margin, we used a linear regression with the residuals of the respective genetic parameters as explanatory variables.
We analyzed population differentiation by calculating overall and pairwise estimates of FST (Weir and Cockerham 1984) among sampling locations, conducting allelic permutation tests for each locus independently (10,000 permutations) in GENEPOP 4.01 (Raymond and Rousset 1995). Additionally to the traditional fixation index (FST) we calculated the diversity index Dest (Jost 2008) using the web based program SMOGD v1.2.5 (Crawford 2010). Similar to FST, Dest is a relative measure of differentiation which ranges from zero (no differentiation) to one (complete differentiation) but contrary to the former, Dest is a more accurate measure when overall divergence is large as it accounts for differences in allele identities. Simulations have shown that Dest outperforms FST over a range of sample sizes and for markers with different numbers of alleles, e.g. for highly variable microsatellite loci (Gerlach et al. 2010).
We conducted Mantel tests (Mantel 1967) in GENEPOP 4.01 (1000 permutations) (Raymond and Rousset 1995) to examine the relationship between the genetic distance measures FST, Dest and the geographic distance between the sampling locations measured as the straight line (Euclidean) distance.
We used two different model-based Bayesian clustering methods, STRUCTURE v.2.2.3 (Pritchard et al. 2000; Falush et al. 2007) and BAPS v5.1 (Corander et al. 2003; Corander and Marttinen 2006) to identify the number of genetically distinct groups in the study region and to assign the sampled individuals to these groups. We first used STRUCTURE v.2.2.3 to test for the most likely number of genetic clusters (K) ranging from 1 to 15, assuming correlated allele frequencies and using sampling localities as prior information (LocPrior). We ran the simulations with a burn-in of 50,000 followed by 100,000 MCMC (Markov Chain Monte Carlo) iterations and with ten runs per K. To identify the most probable number of clusters we used the posterior probability of the data for a given K, LnP(D), and ∆K values as described by Evanno et al. (2005). Following the recommendations of François and Durand (2010) we accounted for admixture and ran spatially explicit models in BAPS v5.1. Because BAPS incorporates geographic information of the samples, we reanalyzed the data in BAPS v5.1 which uses a stochastic optimization algorithm (Corander et al. 2008a) rather than MCMC simulation as in standard Bayesian hierarchical modelling techniques, and automatically estimate the most probable number of genetic clusters (K). The analysis was done in two steps; in the first step we ran a spatial clustering of individuals to allocate each sample into its most likely genetic group using the geographic coordinates of the samples to create a Voronoi tessellation representing spatial population structure (Corander et al. 2008b). We tested values of K ranging from 1 to 15 genetic clusters and ran five iterations for each value of K. The most likely number of clusters was assessed by using changes in the logarithm of the maximum likelihood for each value of K and the Bayesian posterior probability (PP) computed for the ten best partitions (Corander et al. 2006). The PP value closest to 1 identifies the most likely number of genetic clusters supported by the data. In the second step, we used the results obtained from the spatial clustering of individuals to run an admixture model to estimate the proportion of an individual’s ancestry contributed by each of the identified genetic groups (Corander and Marttinen 2006). We used a minimum group size of five individuals to construct admixture sets. Admixture coefficients for individuals were calculated after 50 iterations and following recommendations by Corander and Marttinen (2006), and we used 200 reference individuals to calibrate our estimates.
We tested for signals of recent bottlenecks in the sampled populations, using the statistical approach of Cornuet and Luikart (1996) implemented in the program BOTTLENECK (Piry et al. 1999). The program generates the expected heterozygosity under mutation-drift equilibrium from the number of alleles at a locus and the sample size using different mutation models, and compares it to the average values of observed heterozygosity across all loci and populations. Following the recommendations of Piry et al. (1999), we chose the two phase mutation (TPM) model which applies to most microsatellite loci, with a variance of 30 and a proportion of stepwise mutation model (SMM) in TPM of 70 %. We ran 1000 iterations and due to the small number of loci (<20) we used the Wilcoxon sign-rank test to examine whether the observed heterozygosity deviated from the expected (Piry et al. 1999).
Results
Three loci (Metroe19, MR2-42, and MR3-24) exhibited heterozygote deficiency and deviations from Hardy–Weinberg Equilibrium (p < 0.002; GENEPOP global test) in several populations. This was likely due to the presence of null alleles since deviations were present across all allele sizes. We did not find evidence of linkage disequilibrium among any pairs of loci across all populations (p > 0.05). Of the 29 populations analyzed, only one (Pop 25) showed a significant deviation from HWE (p < 0.006) and an excess of homozygotes (FIS, p < 0.002; Table 1).
Mean allelic richness (AR) per population was 6.92 ± 0.92, ranging from 4.09 to 7.41. Average gene diversity (UHe) across all loci and populations was 0.62 ± 0.05 and varied from 0.48 to 0.68 (Table 1). Populations sampled at the outer margin of the distributional area showed a lower AR, PAR and lower gene diversity (UHe) than the average across all populations, but we found no signs of increased inbreeding (Mann–Whitney U test AR: p = 0.006, PAR: p = 0.023, UHe: p = 0.013 and FIS: p = 0.321 respectively) (Table 1; Fig. 2). Overall, FIS values were low across all sampled populations (mean FIS = 0.032) ranging from −0.089 to 0.136 (Table 1). Analysis of the residuals of the genetic measures showed a positive correlation between distance from the range centre and variability around the mean for all estimates (AR: p = 0.012, UHe: p = 0.003 and FIS: p = 0.009) except for PAR, for which the correlation was negative (PAR: p < 0.001) (Supplementary Fig. 1). Unique alleles (Ua) were mostly found in populations located in the centre of the distribution range. No unique alleles were found in populations at the marginal sites (Table 1; Fig. 1). Although genetic diversity in marginal populations was lower compared to the populations in the centre of the range, no evidence of recent population bottlenecks was found. Analyses of genetic variation among subpopulations using FST and Dest indicated the presence of moderate population differentiation (mean FST = 0.152, SE = 0.052; mean Dest = 0.173 at p < 0.001). Because FST and Dest were showing the same pattern of population differentiation we present only the results of FST (Fig. 3), which is the most commonly used measure in population genetic analyses. The correlation of the geographic (Euclidean) distance between sites and pair-wise FST values of all 29 populations revealed a significant isolation-by-distance pattern (R2 = 0.124, p < 0.0001) (Fig. 3). Interestingly, the data points appear to cluster in two groups.
Bayesian cluster analysis using STRUCTURE indicated the presence of two genetic clusters while BAPS identified ten genetic clusters. Both analyses, STRUCTURE and BAPS, converge on the population structure found in the centre of the range, assigning populations to the same genetic cluster. Additionally, BAPS identified genetic divergence in the marginal populations (Fig. 1). Admixture analyses indicated limited amounts of gene flow among populations (Fig. 4).
Discussion
Contra our first hypothesis, the number of unique alleles and level of allelic richness were not highest in the population at Västerås (Pop 29), which had been the assumed introduction site for M. roeselii in central Sweden based on historical observation records. No unique alleles were found in the Västerås population and genetic diversity was comparable to most parts across the sampled range (Table 1). Instead we found the largest number of unique alleles and highest levels of allelic richness (mean and private allelic richness) at Ullvi (Pop 8) situated 16 km southwest of Västerås. This suggests that M. roeselii rather started expanding from Ullvi and was detected in Västerås at an early stage of the colonization process. The same number of unique alleles but a lower level of allelic richness was found also in a site west of Ullvi (Fellingsbro, Pop 12) which also indicate an early establishment. Interestingly, the sampling site at Ullvi is situated only five km southwest of the royal stud farm of Strömsholm that was established already in the seventeenth century. Strömsholm was a central military riding school and also became a home to the National Swedish Horse Academy. Historical trade records of extensive horse and cattle imports (Bäckström 1924; Montelius 1993) combined with recent genetic analyses (Kaňuch et al. 2013) suggest that M. roeselii was accidentally introduced to this region with hay that arrived with imports from northern Germany or Poland. Species which occupy human modified grassland habitats have an increased likelihood of being introduced to new locations via the transportation of agricultural crops (Wagner 2004). Since M. roeselii overwinters by laying its eggs in hollow grass stems and plant material (Marshall and Haes 1988), we suspect that individuals can be transported with plant materials over large distances. Human mediated dispersal is playing an important role in population expansion and genetic differentiation of other invasive species such as the spread of alien red swamp crayfish in China (Yue et al. 2010), Eurasian dreissenid mussels in USA (Brown and Stepien 2010), and the common wall lizard in Germany (Schulte et al. 2013).
Recurring animal imports from across the Baltic to Västerås and transport to farms in the vicinity appear to have resulted in multiple introductions of individuals as well as the introduction of different lineages of M. roeselii (Kaňuch et al. 2013). This is one possible explanation for the distinct genetic clusters we detected in our analyses. Species dispersal behaviour is also known to affect the genetic structure of populations and the spatiotemporal distribution of genetic variability (Holway and Suarez 1999; Wilson et al. 2009). Metrioptera roeslii is an example of a species with a stratified dispersal pattern, where early colonization and range expansion is driven by rare long-distance dispersal followed by frequent short distance dispersal (“diffusion”) which shapes the population genetic structure (Shigesada et al. 1995; Ibrahim et al. 1996). This is manifested in our data by the two most widespread clusters (shown in yellow and green in Fig. 1) that are not found in well defined regions but rather, to some extent, intermixed. The unique clusters at the range margins also support a pattern that agrees with rare long distance colonisation events.
The overall genetic pattern of the populations sampled around Lake Mälaren appears to reflect the species distribution history (de Jong and Kindvall 1991) and known dispersal biology (Ingrisch and Köhler 1998) very well. Similar pattern of population structure have been observed in other expanding populations in the species native range (Hochkirch and Damerau 2009; Wissmann et al. 2009) and in the UK (Simmons and Thomas 2004). The likelihood that M. roeselii performs long-distance dispersal appears to be low until environmental conditions are favourable and high population densities are reached (Simmons and Thomas 2004; Hochkirch and Damerau 2009; Poniatowski and Fartmann 2009). High propagule pressure and growing population densities in the centre of the species range near Lake Mälaren are likely to have triggered the development of long-winged individuals which were able to colonize distant habitat patches and contribute to the advance of the range margin.
The combination of recurring occasional long distance dispersal by long-winged individuals, frequent short distance movements by short-winged individuals and presumably human aided dispersal at any distance can also explain the genetic pattern of gradually declining genetic variation and increasing variability around the mean towards the range margin. Similar spatiotemporal patterns of genetic diversity have also been observed in other invasive species that rapidly expand their range (Dong et al. 2013, Schulte et al. 2013, Signorile et al. 2014).
The fact that we found low levels of genetic variation in the populations at the range margin suggests that they have been recently founded by a very small number of individuals. A recent study conducted by Kaňuch et al. (2014) confirms our findings that both allelic richness and unbiased expected heterozygosity in isolated populations are correlated with population age. The lack of evidence of recent bottlenecks in any of the sampled populations and only slightly elevated levels of homozygosity indicate that immediate consequences of founder effects are not traceable (Luikart and Cornuet 1998). It is likely that the populations at the range margin have not reached a demographic equilibrium yet and the genetic composition is expected to change with time as a consequence of genetic drift and gene flow (Bohonak 1999). Metrioptera roeseliis ability to regain genetic diversity appears to contribute to the species successful range expansion in northern Europe (Kaňuch et al. 2014; Wissmann et al. 2009).
The moderately decreasing levels of genetic diversity with increasing distance from the core area and evidence of a weak but significant isolation-by-distance pattern in combination with the marginal pockets of low and altered variation are further indications of moderate levels of short distance gene flow distorted by rare long distance dispersal. The weak isolation-by-distance pattern may also suggest a presence of unidentified geographic barriers present in the landscape that restrict the movement of dispersing individuals. The effect of landscape configuration, presence of geographic barriers such as large bodies of water or other types of unsuitable habitat and frequent land use changes causing habitat fragmentation have been shown to affect species genetic structure in other regions (Diekötter et al. 2010, Holzhauer et al. 2006, Lange et al. 2010; Pierce et al. 2014). The landscape around Lake Mälaren consists of a mosaic of agricultural land, forests intersected by small streams, roads and settlements that create a mosaic of more or less permeable habitat and non-habitat. An intensified or altered land use may result in increased fragmentation of landscape that affects species distribution and dispersal.
The majority of M. roeselii individuals in the region around Lake Mälaren are short winged and have a limited movement radius. Despite the occasional occurrence of long-winged individuals and human mediated dispersal, the level of gene flow appears not be high enough to even out variation among populations of M. roeselli as we found a significant differentiation amongst populations. The combination of the species stratified dispersal (species dispersal behaviour and transport by humans) and landscape composition and configuration could be the explanation for the complex spatial genetic structure of M. roeselii around Lake Mälaren. The creation of new grassland habitats within the region can increase dispersal success, and this may result in an increase of genetic diversity and a reduction of genetic differentiation (Gibbs 2001; Lange et al. 2010) as was found in the more central areas around lake Mälaren.
In order to understand in detail what is shaping the genetic population structure and driving the species range expansion further analyses of the exact links between landscape composition, dispersal behaviour and spatiotemporal aspects of genetic variation are needed. Technical and methodological advances will facilitate future genomic data analyses combined with analyses of complex landscape datasets, which will further improve our understanding of the continuous interplay between evolutionary adaptations, genetic drift and homogenizing gene flow during range expansions (Alleaume-Benharira et al. 2006; Kawecki 2008; Rice and Papadopoulos 2009; Bridle et al. 2010).
References
Alleaume-Benharira M, Pen IR, Ronce O (2006) Geographical patterns of adaptation within a species’ range: interactions between drift and gene flow. J Evol Biol 19:203–215
Allendorf FW, Lundquist LL (2003) Introduction: population biology, evolution, and control of invasive species. Conserv Biol 17:24–30
Bäckström A (1924) Strömsholm förr och nu. Författarens Förlag, Stockholm
Bellmann H (2006) Der Kosmos Heuschreckenführer—Die Arten Mitteleuropas sicher bestimmen. Franckh-Kosmos Verlags-Gmbh and Co, Stuttgart
Berggren Å (2004) Impact of grazing on individual male movement in Roesel’s bush-cricket Metrioptera roeseli: one possible clue to species range expansion. J Insect Behav 17:419–429
Berggren A, Low M (2004) Exclusion of the native bog bush-cricket Metrioptera brachyptera by the currently invading Roesel’s bush-cricket Metrioptera roeseli. Entomol Tidskr 125:125–132
Berggren A, Carlson A, Kindvall O (2001) The effect of landscape composition on colonization success, growth rate and dispersal in introduced bush-crickets Metrioptera roeseli. J Anim Ecol 70:663–670
Bialozyt R, Ziegenhagen B, Petit RJ (2006) Contrasting effects of long distance seed dispersal on genetic diversity during range expansion. J Evol Biol 19:12–20
Bohonak A (1999) Dispersal, gene flow, and population structure. Quart Rev Biol 74:21–45
Bridle JR, Polechová J, Kawata M et al (2010) Why is adaptation prevented at ecological margins? New insights from individual-based simulations. Ecol Lett 13:485–494
Brookfield JFY (1996) A simple new method for estimating null allele frequency from heterozygote deficiency. Mol Ecol 5:453–455
Brown J (2004) Tracing the origin of cryptic insect pests and vectors, and their natural enemies. In: Ehler L, Sforza R, Mateille T (eds) Genetics, evolution and biological control. CABI, Oxon, pp 113–135
Brown JE, Stepien CA (2010) Population genetic history of the dreissenid mussel invasions: expansion patterns across North America. Biol Invasions 12:3687–3710
Corander J, Marttinen P (2006) Bayesian identification of admixture events using multilocus molecular markers. Mol Ecol 15:2833–2843
Corander J, Waldmann P, Sillanpaa M (2003) Bayesian analysis of genetic differentiation between populations. Genetics 163:367–374
Corander J, Marttinen P, Mäntyniemi S (2006) Bayesian identification of stock mixtures from molecular marker data. Fish Bull 1004:550–558
Corander J, Marttinen P, Siren J et al (2008a) Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinform 9:539
Corander J, Sirén J, Arjas E (2008b) Bayesian spatial modeling of genetic population structure. Comput Stat 23:111–129
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Crawford NG (2010) SMOGD: software for the measurement of genetic diversity. Mol Ecol Res 10:556–557
de Jong J, Kindvall O (1991) Cikadavårtbitaren Metrioptera roeseli—nykomling eller hotad relikt? (The Roesel’s bush-cricket Metrioptera roeseli—new in Sweden or a threatened relict species?). Fauna och Flora 86:86
Diekötter T, Baveco H, Arens P et al (2010) Patterns of habitat occupancy, genetic variation and predicted movement of a flightless bush cricket, Pholidoptera griseoaptera, in an agricultural mosaic landscape. Landsc Ecol 25:449–461
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Dong et al (2013) Spatial genetic heterogeneity in populations of a newly invasive whitefly in China revealed by a nation-wide field survey. PLoS One. doi:10.1371/journal.pone.0079997
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
François O, Durand E (2010) Spatially explicit Bayesian clustering models in population genetics. Mol Ecol Res 10:773–784
Gardiner T (2009) Macropterism of Roesel’s Bushcricket Metrioptera roeselii in relation to climate change and landscape structure in Eastern England. J Ortho Res 18:95–102
Gerlach G, Jueterbock A, Kraemer P et al (2010) News and views: calculations of population differentiation based on GST and D: forget GST but not all of statistics! Mol Ecol 19:3845–3852
Gibbs JP (2001) Demography versus habitat fragmentation as determinants of genetic variation in wild populations. Biol Conserv 100:15–20
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Updated from Goudet (1995) edn. Available from http://www.unil.ch/izea/softwares/fstat.html
Hochkirch A, Damerau M (2009) Rapid range expansion of a wing-dimorphic bush-cricket after the 2003 climatic anomaly. Biol J Linn Soc 97:118–127
Holway DA, Suarez AV (1999) Animal behavior: an essential component of invasion biology. TREE 14:328–330
Holzhauer SIJ, Wolff K (2005) Polymorphic microsatellite loci in the bush-cricket Metrioptera roeseli. Mol Ecol Notes 5:502–503
Holzhauer S, Ekschmitt K, Sander A-C et al (2006) Effect of historic landscape change on the genetic structure of the bush-cricket Metrioptera roeseli. Landsc Ecol 21:891–899
Holzhauer S, Wolff K, Wolters V (2009) Changes in land use and habitat availability affect the population genetic structure of Metrioptera roeselii (Orthoptera: Tettigoniidae). J Insect Conserv 13:543–552
Hulme PE (2006) Beyond control: wider implications for the management of biological invasions. J Appl Ecol 43:835–847
Ibrahim KM, Nichols RA, Hewitt GM (1996) Spatial patterns of genetic variation generated by different forms of dispersal during range expansion. Heredity 77:282–291
Idar M (1973) Metrioptera roeselii Hbg., en för Sverige ny vårtbitare (Salt. Ensifera). Entomol Tidskrift 94:115
Ingrisch S, Köhler G (1998) Die Heuschrecken Mitteleuropas. Westarp-Wiss, Magdeburg
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Kaňuch P, Pfunder M, Berggren Å et al (2010) Description of nine new microsatellite loci in Metrioptera roeselii (Orthoptera: Tettigoniidae) and their multiplex PCR protocols. Mol Ecol Res 10:404–408
Kaňuch P, Berggren Å, Cassel-Lundhagen A (2013) Colonization history of Metrioptera roeselii in northern Europe indicates human-mediated dispersal. J Biogeogr 40:977–987
Kaňuch P, Berggren Å, Cassel-Lundhagen A (2014) Genetic diversity of a successful colonizer: isolated populations of Metrioptera roeselii regain variation at an unusually rapid rate. Ecol Evol 4:1117–1126
Kawecki TJ (2008) Adaptation to marginal habitats. Ann Rev Ecol Evol Syst 39:321–342
Keller SR, Taylor DR (2010) Genomic admixture increases fitness during a biological invasion. J Evol Biol 23:1720–1731
Kimura M, Weiss GH (1964) The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics 49:561–576
Kindvall O, Vessby K, Berggren Å et al (1998) Individual mobility prevents an Allee effect in sparse populations of the bush cricket Metrioptera roeseli: an experimental study. Oikos 81:449–457
Kirk H, Dorn S, Mazzi D (2013) Molecular genetics and genomics generate new insights into invertebrate pest invasions. Evol Appl 6:842–856
Kleukers R, van Nieukerken E, Ode B et al (1997) De sprinkhanen en krekels van Nederland (Orthoptera)., The grasshoppers and crickets of the Netherlands (Orthoptera)KNNV UitgeveriJ, Utrecht
Lange R, Durka W, Holzhauer SIJ et al (2010) Differential threshold effects of habitat fragmentation on gene flow in two widespread species of bush crickets. Mol Ecol 19:4936–4948
Lawson Handley LJ, Estoup A, Evans DM et al (2011) Ecological genetics of invasive alien species. Biocontrol 56:409–428
Le Corre V, Kremer A (1998) Cumulative effects of founding events during colonisation on genetic diversity and differentiation in an island and stepping-stone model. J Evol Biol 11:495–512
Luikart G, Cornuet J-M (1998) Empirical evaluation of a test for identifying recently bottlenecked populations from Allele frequency data. Conserv Biol 12:228–237
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Marshall J, Haes E (1988) Grasshoppers and allied insects of Great Britain and Ireland. Harley Books, Martins, Great Horkesley, Colchester, Essex
Montelius S (1993) Västerås genom tiderna (In Swedish). Västerås Stad, Västerås
Nichols R, Hewitt G (1994) The genetic consequences of long distance dispersal during colonization. Heredity 72:312–317
Pierce et al. (2014) Serial founder effects and genetic differentiation during worldwide range expansion of monarch butterflies. http://rspb.royalsocietypublishing.org/content/281/1797/20142230
Piry S, Luikart G, Cornuet JM (1999) Computer note. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. J Hered 90:502–503
Poniatowski D, Fartmann T (2009) Experimental evidence for density-determined wing dimorphism in two bush-crickets (Ensifera: Tettigoniidae). Euro J Entomol 106:599–605
Poniatowski D, Fartmann T (2011) Does wing dimorphism affect mobility in Metrioptera roeselii (Orthoptera: Tettigoniidae)? Euro J Entomol 108:409–415
Preuss S, Lundhagen A, Berggren Å (2011) Modelling the distribution of the invasive Roesel’s bush-cricket (Metrioptera roeselii) in a fragmented landscape. NeoBiota 11:33–49
Preuss S, Low M, Cassel-Lundhagen A et al (2014) Evaluating range-expansion models for calculating nonnative species’ expansion rate. Ecol Evol 4:2812–2822
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Puth LM, Post DM (2005) Studying invasion: have we missed the boat? Ecol Lett 8:715–721
Raymond M, Rousset F (1995) GENEPOP (version 1.2): a population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Rice SH, Papadopoulos A (2009) Evolution with stochastic fitness and stochastic migration. PLoS One 4:e7130
Roff DA (1986) The evolution of wing dimorphism in insects. Evolution 40:1009–1020
Ronnås C, Cassel-Lundhagen A, Battisti A et al (2011) Limited emigration from an outbreak of a forest pest insect. Mol Ecol 20:4606–4617
Sakai AK, Allendorf FW, Holt JS et al (2001) The population biology of invasive species. Ann Rev Ecol Syst 32:305–332
Sax D, Stachowicz J, Gaines S (2005) Species invasions: Insights into ecology, evolution, and biogeography. Sinauer Associates, Inc., Sunderland
Schulte U, Veith M, Mingo V et al (2013) Strong genetic differentiation due to multiple founder events during a recent range expansion of an introduced wall lizard population. Biol Invas 15:2639–2649
Shigesada N, Kawasaki K, Takeda Y (1995) Modeling stratified diffusion in biological invasions. Am Midl Nat 146:229–251
Signorile AL, Wang J, Lurz PWW et al (2014) Do founder size, genetic diversity and structure influence rates of expansion of North American grey squirrels in Europe? Div Distr 20:918–930
Simmons A, Thomas C (2004) Changes in dispersal during species’ range expansions. The Am Nat 164:378–395
Suarez AV, Tsutsui ND (2008) The evolutionary consequences of biological invasions. Mol Ecol 17:351–360
Szpiech ZA, Jakobsson M, Rosenberg NA (2008) ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24:2498–2504
Thomas JA, Bourn NAD, Clarke RT et al (2001) The quality and isolation of habitat patches both determine where butterflies persist in fragmented landscapes. Proc R Soc Lond Ser B: Biol Sci 268:1791–1796
Van Oosterhout C, Weetman D, Hutchinson WF (2006) Estimation and adjustment of microsatellite null alleles in nonequilibrium populations. Mol Ecol Notes 6:255–256
Vickery VR (1965) Factors governing the distribution and dispersal of the recently introduced grasshopper, Metrioptera roeseli (Hgb.) (Orthoptera: Ensifera). Ann Entomol Soc Québec 10:165–171
Wagner C (2004) Passive dispersal of Metrioptera bicolor (Phillipi 1830) (Orthopteroidea: Ensifera: Tettigoniidae) by transfer of hay. J Insect Cons 8:287–296
Walsh S, Metzger D, Higuchi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wilson JRU, Dormontt EE, Prentis PJ et al (2009) Something in the way you move: dispersal pathways affect invasion success. TREE 24:136–144
Wissmann J, Schielzeth H, Fartmann T (2009) Landscape-scale expansion of Roesel’s bush-cricket Metrioptera roeselii at the North-western range limit in Central Europe (Orthoptera: Tettigoniidae). Entomologia Generalis 31:317–326
Wright S (1943) Isolation by distance. Genetics 28:114–138
Yue GH, Li J, Bai Z, Wang CM, Feng F (2010) Genetic diversity and population structure of the invasive alien red swamp crayfish. Biol Invasions 12:2697–2706
Acknowledgments
We thank Emilia Broberg and Carin Eriksson for assistance in the field and Johan Wallén for developing the multiplex PCR assays. We are grateful to Bengt Hansson, Tytti Vanhala and Matthew Low for assistance with the data analyses, and JC Ruiz Guajardo and Göran Nordlander for valuable comments on an earlier version of the manuscript. The research was funded by a research grant from Formas to ÅB and Carl Trygger´s foundation to ACL.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Residuals of four genetic measures plotted against geographic distance (km) from the range centre and towards the range edge. There was a positive correlation between distance and variability around the mean for all estimates (AR: p = 0.012, UHe: p = 0.003 and FIS: p = 0.009) except for PAR, for which the correlation was negative (PAR: p < 0.001) (PDF 303 kb)
Supplementary Table 1
Metrioptera roeselii microsatellite markers. Primer names, GenBank accession numbers, fluorescent dyes, PCR multiplex and mean number of alleles corrected for sample size for the 13 microsatellite loci used. Forward (5’-3’) primers were directly labelled with FAM (blue), VIC (green), NED (black) or PET (red) fluorescent dye (DOC 16 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Preuss, S., Berggren, Å. & Cassel-Lundhagen, A. Genetic patterns reveal an old introduction event and dispersal limitations despite rapid distribution expansion. Biol Invasions 17, 2851–2862 (2015). https://doi.org/10.1007/s10530-015-0915-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-015-0915-2