Abstract
Objectives
To search for a novel glutamate decarboxylase (GAD) with an optimum pH towards near-neutrality in order to improve production of gamma-aminobutyric acid (GABA) in recombinant hosts.
Results
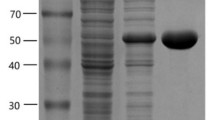
A novel glutamate decarboxylase, BmGAD, from Bacillus megaterium was overexpressed and purified. BmGAD was approximately 53 kDa by SDS-PAGE analysis. Its optimum activity was at pH 5 and 50 °C. BmGAD had a specific activity of 59 ± 5.2 U mg−1 at pH 6, which is the highest value reported so far. The apparent Km and Vmax values of BmGAD were 8 ± 0.5 mM and 150 ± 4.7 U mg−1, respectively. Through site-directed mutagenesis, two BmGAD mutants (E294R and H467A) showed higher Vmax values than that of wild-type, with the values of 210 ± 6.9 and 180 ± 4.1 U mg−1 at pH 5 and 50 °C, respectively.
Conclusions
The unusual high activity of BmGAD at pH 6 makes it an attractive GABA-producing candidate in industrial application.




Similar content being viewed by others
References
Capitani G, De Biase D, Aurizi C, Gut H, Bossa F, Grutter MG (2003) Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J 22:4027–4037
Choi JW, Yim SS, Lee SH, Kang TJ, Park SJ, Jeong KJ (2015) Enhanced production of gamma-aminobutyrate (GABA) in recombinant Corynebacterium glutamicum by expressing glutamate decarboxylase active in expanded pH range. Microb Cell Fact 14:21
Dutyshev DI et al (2005) Structure of Escherichia coli glutamate decarboxylase (GADalpha) in complex with glutarate at 2.05 angstroms resolution. Acta Crystallogr D Biol Crystallogr 61:230–235
Edelheit O, Hanukoglu A, Hanukoglu I (2009) Simple and efficient site-directed mutagenesis using two single-primer reactions in parallel to generate mutants for protein structure-function studies. BMC Biotechnol 9:61
Ge S, Goh EL, Sailor KA, Kitabatake Y, Ming GL, Song H (2006) GABA regulates synaptic integration of newly generated neurons in the adult brain. Nature 439:589–593
Karladee D, Suriyong S (2012) Gamma-Aminobutyric acid (GABA) content in different varieties of brown rice during germination. Scienceasia 38:13–17
Kim HW, Kashima Y, Ishikawa K, Yamano N (2009) Purification and characterization of the first archaeal glutamate decarboxylase from Pyrococcus horikoshii. Biosci Biotechnol Biochem 73:224–227
Park JY, Jeong SJ, Kim JH (2014) Characterization of a glutamate decarboxylase (GAD) gene from Lactobacillus zymae. Biotechnol Lett 36:1791–1799
Pennacchietti E, Lammens TM, Capitani G, Franssen MC, John RA, Bossa F, De Biase D (2009) Mutation of His465 alters the pH-dependent spectroscopic properties of Escherichia coli glutamate decarboxylase and broadens the range of its activity toward more alkaline pH. J Biol Chem 284:31587–31596
Seo MJ, Nam YD, Lee SY, Park SL, Yi SH, Lim SI (2013) Expression and characterization of a glutamate decarboxylase from Lactobacillus brevis 877G producing gamma-aminobutyric acid. Biosci Biotechnol Biochem 77:853–856
Shi F, Xie Y, Jiang J, Wang N, Li Y, Wang X (2014) Directed evolution and mutagenesis of glutamate decarboxylase from Lactobacillus brevis Lb85 to broaden the range of its activity toward a near-neutral pH. Enzyme Microb Technol 61–62:35–43
Thu Ho NA, Hou CY, Kim WH, Kang TJ (2013) Expanding the active pH range of Escherichia coli glutamate decarboxylase by breaking the cooperativeness. J Biosci Bioeng 115:154–158
Wang Q, Xin Y, Zhang F, Feng Z, Fu J, Luo L, Yin Z (2011) Enhanced γ-aminobutyric acid-forming activity of recombinant glutamate decarboxylase (gadA) from Escherichia coli. World J Microb Biot 27:693–700
Wang N, Ni Y, Shi F (2015) Deletion of odhA or pyc improves production of gamma-aminobutyric acid and its precursor L-glutamate in recombinant Corynebacterium glutamicum. Biotechnol Lett 37:1473–1481
Yu K, Lin L, Hu S, Huang J, Mei L (2012) C-terminal truncation of glutamate decarboxylase from Lactobacillus brevis CGMCC 1306 extends its activity toward near-neutral pH. Enzyme Microb Technol 50:263–269
Zhang C, Lu J, Chen L, Lu F, Lu Z (2014) Biosynthesis of γ-aminobutyric acid by a recombinant Bacillus subtilis strain expressing the glutamate decarboxylase gene derived from Streptococcus salivarius ssp. thermophilus Y2. Proc Biochem 49:1851–1857
Acknowledgments
This research was supported by “Hundred Talents Program” of the Chinese Academy of Sciences, the National Natural Science Foundation of China (No. 31500044, 31571029, 31501475) and the Natural Science Foundation of Tianjin (15JCYBJC30300, 15JCTPJC55400).
Supporting Information
Supplementary Table 1 Strains and plasmids used in this study.
Supplementary Table 2 Primers used in this study.
Supplementary Fig. 1 Enzymatic transformation of glutamate to GABA at near-neutral pH.
Supplementary Fig. 2 Determination of kinetic parameters for the recombinant BmGAD.
Supplementary Fig. 3 Multiple alignments of B. megaterium GAD with other reported GADs.
Author information
Authors and Affiliations
Corresponding author
Additional information
Qingdai Liu and Haijiao Cheng have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, Q., Cheng, H., Ma, X. et al. Expression, characterization and mutagenesis of a novel glutamate decarboxylase from Bacillus megaterium . Biotechnol Lett 38, 1107–1113 (2016). https://doi.org/10.1007/s10529-016-2070-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2070-y