Abstract
Objectives
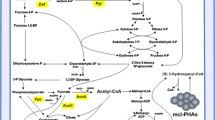
To enhance the biosynthesis of medium-chain-length polyhydroxyalkanoates (PHAMCL) from glucose in Pseudomonas mendocina NK-01, metabolic engineering strategies were used to block or enhance related pathways.
Results
Pseudomonas mendocina NK-01 produces PHAMCL from glucose. Besides the alginate oligosaccharide biosynthetic pathway proved by our previous study, UDP-d-glucose and dTDP-l-rhamnose biosynthetic pathways were identified. These might compete for glucose with the PHAMCL biosynthesis. First, the alg operon, galU and rmlC gene were deleted one by one, resulting in NK-U-1(∆alg), NK-U-2 (∆alg∆galU), NK-U-3(alg∆galU∆rmlC). After fermentation for 36 h, the cell dry weight (CDW) and PHAMCL production of these strains were determined. Compared with NK-U: 1) NK-U-1 produced elevated CDW (from 3.19 ± 0.16 to 3.5 ± 0.11 g/l) and equal PHAMCL (from 0.78 ± 0.06 to 0.79 ± 0.07 g/l); 2) NK-U-2 produced more CDW (from 3.19 ± 0.16 to 3.55 ± 0.23 g/l) and PHAMCL (from 0.78 ± 0.06 to 1.05 ± 0.07 g/l); 3) CDW and PHAMCL dramatically decreased in NK-U-3 (1.53 ± 0.21 and 0.41 ± 0.09 g/l, respectively). Additionally, the phaG gene was overexpressed in strain NK-U-2. Although CDW of NK-U-2/phaG decreased to 1.29 ± 0.2 g/l, PHA titer (%CDW) significantly increased from 24.5 % up to 51.2 %.
Conclusion
The PHAMCL biosynthetic pathway was enhanced by blocking branched metabolic pathways in combination with overexpressing phaG gene.



Similar content being viewed by others
References
Borrero-de Acuna JM et al (2014) Production of medium chain length polyhydroxyalkanoate in metabolic flux optimized Pseudomonas putida. Microb Cell Fact 13:88
Byrd MS et al (2009) Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol 73:622–638
de Eugenio LI, Escapa IF, Morales V, Dinjaski N, Galan B, Garcia JL, Prieto MA (2010) The turnover of medium-chain-length polyhydroxyalkanoates in Pseudomonas putida KT2442 and the fundamental role of PhaZ depolymerase for the metabolic balance. Environ Microbiol 12:207–221
Escapa IF, Morales V, Martino VP, Pollet E, Averous L, Garcia JL, Prieto MA (2011) Disruption of beta-oxidation pathway in Pseudomonas putida KT2442 to produce new functionalized PHAs with thioester groups. Appl Microbiol Biot 89:1583–1598
Guo W, Song C, Kong M, Geng W, Wang Y, Wang S (2011a) Simultaneous production and characterization of medium-chain-length polyhydroxyalkanoates and alginate oligosaccharides by Pseudomonas mendocina NK-01. Appl Microbiol Biotechnol 92:791–801
Guo W et al (2011b) Complete genome of Pseudomonas mendocina NK-01, which synthesizes medium-chain-length polyhydroxyalkanoates and alginate oligosaccharides. J Bacteriol 193:3413–3414
Guo W, Feng J, Geng W, Song C, Wang Y, Chen N, Wang S (2012) Augmented production of alginate oligosaccharides by the Pseudomonas mendocina NK-01 mutant. Carbohyd Res 352:109–116
Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP (1998) A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86
Jendrossek D, Pfeiffer D (2014) New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ Microbiol 16:2357–2373
Leong YK, Show PL, Ooi CW, Ling TC, Lan JC (2014) Current trends in polyhydroxyalkanoates (PHAs) biosynthesis: insights from the recombinant Escherichia coli. J Biotechnol 180:52–65
Liu Q, Luo G, Zhou XR, Chen GQ (2011) Biosynthesis of poly(3-hydroxydecanoate) and 3-hydroxydodecanoate dominating polyhydroxyalkanoates by beta-oxidation pathway inhibited Pseudomonas putida. Metab Eng 13:11–17
Ouyang SP, Luo RC, Chen SS, Liu Q, Chung A, Wu Q, Chen GQ (2007) Production of polyhydroxyalkanoates with high 3-hydroxydodecanoate monomer content by fadB and fadA knockout mutant of Pseudomonas putida KT2442. Biomacromolecules 8:2504–2511
Poblete-Castro I et al (2012) The metabolic response of P. putida KT2442 producing high levels of polyhydroxyalkanoate under single- and multiple-nutrient-limited growth: highlights from a multi-level omics approach. Microb Cell Fact 11:34
Poblete-Castro I, Binger D, Rodrigues A, Becker J, Martins Dos Santos VA, Wittmann C (2013) In-silico-driven metabolic engineering of Pseudomonas putida for enhanced production of poly-hydroxyalkanoates. Metab Eng 15:113–123
Qiu YZ, Han J, Guo JJ, Chen GQ (2005) Production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from gluconate and glucose by recombinant Aeromonas hydrophila and Pseudomonas putida. Biotechnol Lett 27:1381–1386
Wang Q, Tappel RC, Zhu C, Nomura CT (2012) Development of a new strategy for production of medium-chain-length polyhydroxyalkanoates by recombinant Escherichia coli via inexpensive non-fatty acid feedstocks. Appl Environ Microb 78:519–527
Wang Y et al (2015) An upp-based markerless gene replacement method for genome reduction and metabolic pathway engineering in Pseudomonas mendocina NK-01 and Pseudomonas putida KT2440. J Microbiol Methods 113:27–33
Acknowledgments
This work was supported by the National Key Basic Research Program of China (“973”-Program) 2012CB725204, the National Key Technology Support Program 2015BAD16B04, the Natural Science Foundation of China Grant Nos. 31470213 and 31170030, and the Project of Tianjin, China (13JCZDJC27800, 13JCYBJC24900, 13TXSYJC40100 and 14ZCZDSF00009).
Supporting information
Supplementary Table 1—Primers used in this study.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Y., Zhao, F., Fan, X. et al. Enhancement of medium-chain-length polyhydroxyalkanoates biosynthesis from glucose by metabolic engineering in Pseudomonas mendocina . Biotechnol Lett 38, 313–320 (2016). https://doi.org/10.1007/s10529-015-1980-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-015-1980-4