Abstract
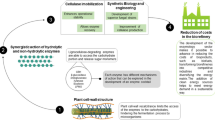
The complex structure of lignocellulose requires the involvement of a suite of lignocellulolytic enzymes for bringing about an effective de-polymerization. Cellulases and hemicellulases from both fungi and bacteria have been studied extensively. This review illustrates the mechanism of action of different cellulolytic and hemi-cellulolytic enzymes and their distinctive roles during hydrolysis. It also examines how different approaches can be used to improve the synergistic interaction between fungal and bacterial glycosyl hydrolases with a focus on fungal cellulases and bacterial hemicellulases. The approach entails the role of cellulosomes and their improvement through incorporation of novel enzymes and evaluates the recent break-through in the construction of designer cellulosomes and their extension towards improving fungal and bacterial synergy. The proposed approach also advocates the incorporation and cell surface display of designer cellulosomes on non-cellulolytic solventogenic strains along with the innovative application of combined cross-linked enzyme aggregates (combi-CLEAs) as an economically feasible and versatile tool for improving the synergistic interaction through one-pot cascade reactions.

Similar content being viewed by others
References
Adav SS, Chao LT, Sze SK (2012) Quantitative secretomic analysis of Trichoderma reesei strains reveals enzymatic composition for lignocellulosic biomass degradation. Mol Proteomics 11(7):M111.012419. doi:10.1074/mcp.M111,012419
Arfia Y, Shamshouma M, Rogachevb I et al (2014) Integration of bacterial lytic polysaccharide monooxygenases into designer cellulosomes promotes enhanced cellulose degradation. Proc Natl Acad Sci USA 111:9109–9114
Aro N, Pakula T, Penttila M (2005) Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol Rev 29:719–739
Baker JO, Adney WS, Thomas SR et al (1995) Synergism between purified bacterial and fungal cellulases. Enzym Degrad Ins Carb 618:113–141
Baker JO, Ehrman CI, Adney WS et al (1998) Hydrolysis of cellulose using ternary mixtures of purified celluloses. Appl Biochem Biotech 70–72:395–403
Bayer EA, Lamed R, Himmel ME (2007) The potential of cellulases and cellulosomes for cellulosic waste management. Curr Opin Biotechnol 18:237–245
Beckham GT, Matthews JF, Bomble YJ et al (2010) Identification of amino acids responsible for processivity in a family 1 carbohydrate-binding module from a fungal cellulase. J Phys Chem B 114:1447–1453
Bhattacharya A, Pletschke BI (2014) Magnetic cross-linked enzyme aggregates (CLEAs): a novel concept towards carrier free immobilization of lignocellulolytic enzymes. Enz Microb Technol 61–62:17–27
Bhattacharya A, Shrivastava A, Sharma A (2013) Evaluation of enhanced thermostability and operational stability of carbonic anhydrase from Micrococcus species. Appl Biochem Biotechnol 170:756–763
Binod P, Janu KU, Sindhu R et al (2011) Hydrolysis of lignocellulosic biomass for bioethanol production. In: Pandey A, Larroche C, Ricke SC (eds) Biofuels: alternative feedstocks and conversion processes. Elsevier Inc., Amsterdam, pp 229–250
Boraston AB, Bolam DN, Gilbert HJ et al (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382:769–781
Brunecky R, Alahuta M, Xu Q (2013) Revealing nature’s cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science 342:1513
Carrard G, Koivula A, Soderlund H et al (2000) Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc Natl Acad Sci USA 97:10342–10347
Costaouec TL, Pakarinen A, Varnai A et al (2013) The role of carbohydrate binding module (CBM) at high substrate consistency: Comparison of Trichoderma reesei and Thermoascus aurantiacus Cel7A (CBHI) and Cel5A (EGII). Bioresour Technol 143:196–203
Dalal S, Sharma A, Gupta MN (2007) A multipurpose immobilized biocatalyst with pectinase, xylanase and cellulase activities. Chem Cent J 1:1–5
Dashtban M, Schraft H, Qin W (2009) Fungal bioconversion of lignocellulosic residues; opportunities and perspectives. Int J Biol Sci 5:578–595
Den Haan R, Van Zyl WH (2003) Enhanced xylan degradation and utilisation by Pichia stipitis overproducing fungal xylanolytic enzymes. Enz Microb Technol 33:620–662
Dhawan S, Kaur J (2007) Microbial mannanases: an overview of production and applications. Crit Rev Biotechnol 27:197–216
Do TT, Quyen DT, Nguyen TN et al (2013) Molecular characterization of a glycosyl hydrolase family 10 xylanase from Aspergillus niger. Prot Expr Purif 92:196–202
Doi RH, Kosugi A (2004) Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat Rev Microbiol 202:541–551
Doi RH, Kosugi A, Murashima K et al (2003) Cellulosomes from mesophilic bacteria. J Bacteriol 185:5907–5914
Elkins JG, Raman B, Keller M (2010) Engineered microbial systems for enhanced conversion of lignocellulosic biomass. Curr Opin Biotechnol 21:657–662
Fierobe H-P, Bayer EA, Tardif C et al (2002) Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J Biol Chem 277:49621–49630
Fierobe H-P, Mingardon F, Mechaly A et al (2005) Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J Biol Chem 280:16325–16334
Gao D, Chundawat SPS, Krishnan C et al (2010) Mixture optimization of six core glycosyl hydrolases for maximizing saccharification of ammonia fiber expansion (AFEX) pretreated corn stover. Bioresour Technol 101:2770–2781
Gao D, Uppugundla N, Chundawat SPS et al (2011) Hemicellulases and auxiliary enzymes for improved conversion of lignocellulosic biomass to monosaccharides. Biotechnol Biofuel 4:5
Gefen G, Anbar M, Morag E et al (2012) Enhanced cellulose degradation by targeted integration of a cohesin-fused β-glucosidase into the Clostridium thermocellum cellulosome. Proc Natl Acad Sci USA 109:10298–10303
Grun CH, Dekker N, Nieuwland AA et al (2006) Mechanism of action of the endo-(1-3)-α-glucanase MutAp from the mycoparasitic fungus Trichoderma harzianum. FEBS Lett 580:3780–3786
Han Y, Kye C, Hong S (2006) A cel44C-man26A gene of endophytic Paenibacillus polymyxa GS01 has multi-glycosyl hydrolases in two catalytic domains. Appl Microbiol Biotechnol 73:618–630
Hasper AA, Dekkers E, Mil M et al (2002) EglC a new endoglucanase from Aspergillus niger with major activity towards xyloglucan. Appl Environ Microbiol 68:1556–1560
Hatada Y, Takeda N, Hirasawa K et al (2005) Sequence of the gene for a high-alkaline mannanase from an alkaliphilic Bacillus sp. strain JAMB-750, its expression in B. subtilis and characterization of the recombinant enzyme. Extremophiles 9:497–500
Himmel ME, Ding SY, Johnson DK et al (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–809
Hogg D, Pell G, Dupree et al (2003) The modular architecture of Cellvibrio japonicus mannanases in glycoside hydrolase families 5 and 26 points to differences in their role in mannan degradation. Biochem J 371:1027–1043
Horn SJ, Vaaje-Kolstad G, Westereng B et al (2012) Novel enzymes for the degradation of cellulose. Biotechnol Biofuel 5:45
Hu J, Arantes V, Saddler JN (2011) The enhancement of enzymatic hydrolysis of lignocellulosic substrates by the addition of accessory enzymes such as xylanase: is it an additive or synergistic effect? Biotechnol Biofuel 4:36
Irwin DC, Spezio M, Walker LP, Wilson DB (1993) Activity studies of eight purified cellulases: specificity, synergism and binding domain effects. Biotechnol Bioeng 42:1002–1013
Jeon Eugene, Hyeon Jeong-eun, Suh Dong Jin et al (2009) Production of cellulosic ethanol in Saccharomyces cerevisiae heterologous expressing Clostridium thermocellum endoglucanase and Saccharomycopsis fibuligera β-glucosidase genes. Mol Cell 28:369–373
Johnson EA, Sakajoh M, Halliwell G, Madia A, Demain AL (1982) Saccharification of complex cellulosic substrates by the cellulase system from Clostridium thermocellum. Appl Env Microbiol 43:1125–1132
Juturu V, Wu JC (2012) Microbial xylanases. Engineering, production and applications. Biotechnol Adv 30:1219–1227
Kosugi A, Murashima K (2002) Doi RH Xylanase and acetyl xylan esterase activities of XynA, a key subunit of the Clostridium cellulovorans cellulosome for xylan degradation. Appl Environ Microbiol 68:6399–6402
Kovacs K, Willson BJ, Schwarz K (2013) Secretion and assembly of functional mini cellulosomes from synthetic chromosomal operons in Clostridium acetobutylicum ATCC 824. Biotechnol Biofuel 6:117
Kubicek CP, Mikus M, Schuster A et al (2009) Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecornia. Biotechnol Biofuels 2:19
La Grange DC, den Haan R, Willem H et al (2010) Engineering cellulolytic ability into bioprocessing organisms. Appl Microbiol Biotechnol 87:1195–1208
Lairson LL, Henrissat B, Davies GJ et al (2008) Glycosyltransferases: structures functions and mechanisms. Ann Rev Biochem 77:521–555
Lee HL, Chang CK, Jeng WY et al (2012) Mutations in the substrate entrance region of β-glucosidase from Trichoderma reesei improve enzyme activity and thermostability. Protein Eng Des Sel. doi:10.1093/protein/gzs073
Li DC, Li AN, Papageorgiou AC (2011) Cellulases from thermophilic fungi recent insights and biotechnological potential. Enzym Res. doi:10.4061/2011/308730
Liu T, Yun H, Zhang AC et al (2012) Aspergillus niger DLFCC-90 rhamnoside hydrolase, a new type of flavonoid glycoside hydrolase. Appl Environ Microbiol 78:4752–4754
Ljungdahl LG (2008) The cellulase/hemicellulase system of the anaerobic fungus Orpinomyces PC-2 and aspects of its applied use. Ann NY Acad Sci 1125:308–321
Lombard V, Golaconda Ramalu H, Drula E et al (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acid Res 42:D490–D495
Lynd LR, Weimer PJ, van Zyl WH et al (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–577
Maki M, Leung KT, Qin W (2009) The prospects of cellulase producing bacteria for the bioconversion of lignocellulosic biomass. Int J Biol Sci 5:500–516
Martınez D, Berka RM, Henrissat B et al (2008) Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat Biotechnol 26:513–560
Master ER, Zheng Y, Storms R et al (2008) A xyloglucan-specific family 12 glycosyl hydrolase from Aspergillus niger recombinant expression, purification and characterization. Biochem J 411:161–170
Mingardon F, Chanal A, Lopez-Contreras AM et al (2007) Incorporation of fungal cellulases in bacterial minicellulosomes yields viable, synergistically acting cellulolytic complexes. Appl Environ Microbiol 73:3822–3832
Mohanram S, Amat D, Choudhary J et al (2013) Novel perspectives for evolving enzyme cocktails for lignocellulose hydrolysis in biorefineries. Sustain Chem Proc 1:15
Nascimento AS, Muniz JRC, Aparıcio R et al (2014) Insights into the structure and function of fungal β-mannosidases from glycoside hydrolase family 2 based on multiple crystal structures of the Trichoderma harzianum enzyme. FEBS J 281:4165–4178
Olson DG, Tripathi SA, Richard J et al (2010) Deletion of the Cel48S cellulase from Clostridium thermocellum. Proc Natl Acad Sci USA 107:17727–17732
Paes G, Berrin J-G, Beaugrand J (2012) GH11 xylanases: Structure/function/properties relationships and applications. Biotechnol Adv 30:564–592
Pham Hoa PT, Quyen DT et al (2011) Cloning, expression, purification, and properties of an endoglucanase gene (glycosyl hydrolase family 12) from Aspergillus niger VTCC-F021 in Pichia pastoris. J Microbiol Biotechnol 21:1012–1020
Piotek M, Hagedorn J, Hollenberg CP et al (1998) Two novel gene expression systems based on the yeasts Schwanniomyces occidentalis and Pichia stipitis. Appl Microbiol Biotechnol 50:331–338
Prassad S, Singh A, Joshi HC (2007) Ethanol as an alternative fuel from agricultural, industrial and urban residues. Resour Conserv Recycl 50:1–39
Prates JA, Tarbouriech N, Charnock SJ et al (2001) The structure of the feruloyl esterase module of xylanase 10B from Clostridium thermocellum provides insights into substrate recognition. Structure 9:1183–1190
Rabinovich ML, Melnick MS, Bolobova AV (2002) The structure and mechanism of action of cellulolytic enzymes. Biochemistry 67:1026–1050
Rakotoarivonina H, Hermant B, Monthe N et al (2012) The hemicellulolytic enzyme arsenal of Thermobacillus xylanilyticus depends on the composition of biomass used for growth. Microb Cell Fact 11:159
Rojas AL, Fischer H, Elena V et al (2005) Structural insights into the α-xylosidase from Trichoderma reesei obtained by synchrotron small-angle X-ray scattering and circular dichroism spectroscopy. Biochemistry 44:15578–15584
Sanchez C (2009) Lignocellulosic residues: biodegradation and bioconversion by fungi. Biotechnol Adv 27:185–194
Sapag A, Wouters J, Lambert C et al (2002) The endoxylanases from family II: computer analysis of protein sequences reveals important structural and phylogenetic relationships. J Biotechnol 95:109–131
Sethi A, Scharf ME (2013) Biofuels: fungal, bacterial and insect degraders of lignocellulose. eLS. doi:10.1002/9780470015902.a0020374. John Wiley and Sons
Shallom D, Shoham Y (2003) Microbial hemicellulases. Curr Opin Microbiol 6:219–228
Shoseyov O, Shani Z, Levy I (2006) Carbohydrate binding modules: biochemical properties and novel applications. Microbiol Mol Biol Rev 70:283–295
Shrivastava S, Shukla P, Poddar (2007) In silico studies for evaluating conservation homology among family 11 xylanases from Thermomyces lanuginosus. J Appl Sci Environ Sanit 2:70–76
Stalbrand H, Saloheimo A, Vehmaanpera J et al (1995) Cloning and expression in Saccharomyces cerevisiae of a Trichoderma reesei β-mannanase gene containing a cellulose binding domain. Appl Environ Microbiol 61:1090–1097
Stoll D, Boraston A, Stalbrand H et al (2000) Mannanase Man26A from Cellulomonas fimi has a mannan-binding module. FEMS Lett 183:265–269
Subramaniyan S, Prema P (2002) Biotechnology of microbial xylanases: enzymology, molecular biology and application. Crit Rev Biotechnol 22:33–46
Sweeney MD, Xu F (2012) Biomass converting enzymes as industrial biocatalysts for fuels and chemicals: recent developments. Catalysts 2:244–263
Talekar S, Joshi G, Joshi A et al (2013) Parameters in preparation and characterization of cross linked enzyme aggregates (CLEAs). RSC Adv 3:12485
Tang CM, Waterman LD, Smith MH et al (2001) The cel4 gene of Agaricus bisporus encodes a β-mannanase. Appl Environ Microbiol 67:2298–2303
Van den Brink J, De Vries RP (2011) Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol 91:1477–1492
Van Dyk JS, Pletschke BI (2012) A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes-factors affecting enzymes, conversion and synergy. Biotechnol Adv 30:1458–1480
Van Dyk JS, Sakkab M, Sakkab K et al (2010) Identification of endoglucanases, xylanases, pectinases and mannanases in the multi-enzyme complex of Bacillus licheniformis SVD1. Enz Microb Technol 47:112–118
Vikari L, Vehmaanpera J, Koivula A (2012) Lignocellulosic ethanol: from science to industry. Biomass Bioenergy 46:13–24
Wen F, Sun J, Zhoa H (2010) Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Appl Environ Microbiol 76:1251–1260
Wilson DB (2011) Microbial diversity of cellulose hydrolysis. Curr Opin Microbiol 14:1–5
Zhang Q, Yan X, Zhang L et al (2006) Cloning, sequence analysis, and heterologous expression of a β-mannanase gene from Bacillus subtilis Z-2. Mol Biol 40:368–374
Zhengqiang J, Kobayashi A, Ahsan MM et al (2001) Characterization of a thermostable family 10 endoxylanase (XynB) from Thermotoga maritima that cleaves p-nitrophenylbeta-D-xyloside. J Biosci Bioeng 92:423–428
Zverlov VV, Schantz N, Schmitt-Kopplin P et al (2005) Two new major subunits in the cellulosome of Clostridium thermocellum: xyloglucanase Xgh74A and endoxylanase Xyn10D. Microbiology 151:3395–3401
Acknowledgments
Ankita Shrivastava Bhattacharya and Abhishek Bhattacharya are grateful to the National Research Foundation (NRF) of South Africa for their NRF Free Standing Post Doctoral Fellowship awards. Any opinion, findings and conclusions or recommendations expressed in this material are those of the author(s) and therefore the NRF does not accept any liability in regard thereto.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bhattacharya, A.S., Bhattacharya, A. & Pletschke, B.I. Synergism of fungal and bacterial cellulases and hemicellulases: a novel perspective for enhanced bio-ethanol production. Biotechnol Lett 37, 1117–1129 (2015). https://doi.org/10.1007/s10529-015-1779-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-015-1779-3