Abstract
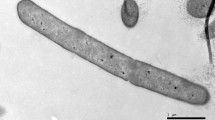
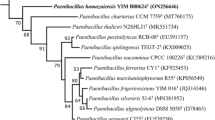
A novel facultatively anaerobic bacterial strain, designated LAM0504T, was isolated from a pit mud of Luzhou flavour liquor alcohol fermentation in Sichuan Province, China. Cells of strain LAM0504T were observed to be Gram-stain negative, spore-forming, rod shaped and motile by means of peritrichous flagella. Strain LAM0504T was found to be able to grow at 20–48 °C (optimum: 30 °C), pH 5.0–9.0 (optimum: 7.0) and 0–3 % NaCl (w/v) (optimum: 1.0 %). The 16S rRNA gene sequence similarity analysis showed that strain LAM0504T was most closely related to Paenibacillus konsisdensis JCM 14798T, Fontibacillus phaseoli LMG 27589T and Paenibacillus motobuensis JCM 12774T, with 97.0, 96.8 and 96.7 % sequence similarity, respectively. The DNA–DNA hybridization value between strain LAM0504T and P. konsisdensis JCM 14798T was 53.3 ± 1.2 %. The genomic DNA G+C content of strain LAM0504T was 43.0 mol% as determined by the Tm method. The major fatty acids of strain LAM0504T were identified as anteiso-C15:0, C16:0 and iso-C15:0. The cell-wall peptidoglycan was found to contain meso-diaminopimelic acid. The predominant menaquinone was identified as MK-7. The major polar lipids were found to be diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, two unidentified phospholipids, two unidentified glycolipids and three unidentified lipids. On the basis of its physiological and phylogenetic characteristics, strain LAM0504T is concluded to represent a novel species of the genus Paenibacillus, for which the name Paenibacillus vini sp. nov. is proposed. The type strain is LAM0504T (=ACCC 06420T = JCM 19842T).

Similar content being viewed by others
References
Ash C, Priest FG, Collins MD (1993) Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek 64:253–260
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Fang M-X, Zhang W-W, Zhang Y-Z, Tan H-Q, Zhang X-Q, Wu M, Zhu X-F (2012) Brassicibacter mesophilus gen. nov., sp. nov., a strictly anaerobic bacterium isolated from food industry wastewater. Int J Syst Evol Microbiol 62:3018–3023
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Flores-Félix JD, Mulas R, Ramírez-Bahena M-H, Cuesta MJ, Rivas R, Barañas J, Mulas D, González-Andrés F, Peix A, Velázquez E (2014) Fontibacillus phaseoli sp. nov. isolated from Phaseolus vulgaris nodules. Antonie Van Leeuwenhoek 105:23–28
Huss VAR, Festl H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184–192
Iida K, Ueda Y, Kawamura Y, Ezaki T, Takade A, Yoshida S, Amako K (2005) Paenibacillus motobuensis sp. nov., isolated from a composting machine utilizing soil from Motobu-town, Okinawa, Japan. Int J Syst Evol Microbiol 55:1811–1816
Kates M (1986) Techniques of lipidology, 2nd edn. Elsevier, Amsterdam
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Ko KS, Kim YS, Lee MY, Shin SY, Jung DS, Peck KR, Song JH (2008) Paenibacillus konsidensis sp. nov., isolated from a patient. Int J Syst Evol Microbiol 58:2164–2168
Komagata K, Suzuki K (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Logan NA, Berge O, Bishop AH, Busse H-L, Vos PD, Fritze D, Heyndrickx M, Kämper P, Rabinovitch L, Salkinoja-Salonen MS, Seldin L, Ventosa A (2009) Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int J Syst Evol Microbiol 59:2114–2121
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218
Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118
Minnikin DE, Odonnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Montes MJ, Mercade E, Bozal N, Guinea J (2004) Paenibacillus antarcticus sp. nov., a novel psychrotolerant organism from the Antarctic environment. Int J Syst Evol Microbiol 54:1521–1526
Osman S, Satomi M, Venkateswaran K (2006) Paenibacillus pasadenensis sp. nov. and Paenibacillus barengoltzii sp. nov., isolated from a spacecraft assembly facility. Int J Syst Evol Microbiol 56:1509–1514
Priest FG (2009) Genus I. Paenibacillus Ash, Priest and Collins 1994, 852VP. In: De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman WB (eds) Bergey’s manual of systematic bacteriology, vol 3, 2nd edn. Springer, New York, pp 269–295
Ruan Z, Wang Y, Song J, Jiang S, Wang H, Li Y, Zhao B, Jiang R, Zhao B (2014) Kurthia huakuii sp. nov., isolated from biogas slurry, and emended description of the genus Kurthia. Int J Syst Evol Microbiol 64:518–521
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sakamoto M, Suzuki M, Umeda M, Ishikawa I, Benno Y (2002) Reclassification of bacteroides forsythus (Tanner et al. 1986) as Tannerella forsythensis corrig., gen. nov., comb. Nov. Int J Syst Evol Microbiol 52:841–849
Schleifer KH (1985) Analysis of the chemical composition and primary structure of murein. Methods Microbiol 18:123–156
Shida O, Takagi H, Kadowaki K, Nakamura LK, Komagata K (1997) Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int J Syst Bacteriol 47:289–298
Simbert RM, Krieg NR (1981) General characterization. In: Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Philips GB (eds) Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC, pp 409–443
Simbert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhare P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 647–654
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Traiwan J, Park M-K, Kim W (2011) Paenibacillus puldeungensis sp. nov., isolated from a grassy sandbank. Int J Syst Evol Microbiol 61:670–673
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Xu X-W, Huo Y-Y, Wang C-S, Oren A, Cui H-L, Vedler E, Wu M (2011) Pelagibacterium halotolerans gen. nov., sp. nov. and Pelagibacterium luteolum sp. nov., novel members of the family Hyphomicrobiaceae. Int J Syst Evol Microbiol 61:1817–1822
Zhou Y, Gao S, Wei DQ, Yang LL, Huang X, He J, Zhang YJ, Tang SK, Li WJ (2012) Paenibacillus thermophilus sp. nov., a novel bacterium isolated from a sediment of hot spring in Fujian province, China. Antonie Van Leeuwenhoek 102:601–609
Acknowledgments
We would like to thank Professor Aharon Oren from The Hebrew University of Jerusalem for assistance with Latin in deriving the specific epithet for strain LAM0504T. We also thank Professor Min Wu from Zhejiang University for his technique assistant on the chemotaxonomic analyses. This work was supported by National Nonprofit Institute Research Grant of CAAS (No. 2014-30), Fund for Innovation team project of environmental pollution preventing and controlling technology (No. 670100656), National Key Technology R&D Program of China (Nos. 2013BAD05B04F02 and 2011BAD11B05), Foundation of the Key Laboratory of Development and Application of Rural Renewable Energy (MOA, China) (No. 2013002), and the Science Foundation of Modern Farming Group (No. MF20100518).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, XR., Shao, CB., Wang, YW. et al. Paenibacillus vini sp. nov., isolated from alcohol fermentation pit mud in Sichuan Province, China. Antonie van Leeuwenhoek 107, 1429–1436 (2015). https://doi.org/10.1007/s10482-015-0438-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-015-0438-y