Abstract
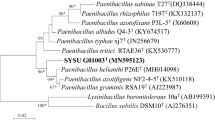
A Gram-positive, thermophilic, strictly aerobic bacterium, designated WP-1T, was isolated from a sediment sample from a hot spring in Fujian province of China and subjected to a polyphasic taxonomic study. Cells of strain WP-1T were rods (~0.6–0.8 × 2.5–3.5 μm) and motile by means of peritrichous flagella. Endospores were ellipsoidal in terminal or subterminal positions. Strain WP-1T grew at 37–60 °C (optimum 42–45 °C), 0–3 % NaCl (optimum 1 %, w/v) and pH 3.0–9.0 (optimum pH 6.5–7.0). The predominant menaquinone was MK-7. The major fatty acids were anteiso-C15:0, iso-C16:0, C16:0 and anteiso-C17:0. The polar lipids consisted of diphosphatidylglycerol, phosphatidylglycerol, two glycolipids, two unidentified phospholipids and two unknown polar lipids. The cell-wall peptidoglycan contained meso-diaminopimelic acid (meso-DAP). The G + C content of the genomic DNA was 52.5 %. Phylogenetic analysis based on the 16S rRNA gene sequence showed that strain WP-1T is a member of the genus Paenibacillus and exhibited sequence similarity of 99.3 % to Paenibacillus macerans DSM 24T and both strains represented a separate lineage from all other Paenibacillus species. However, the level of DNA–DNA relatedness between strain WP-1T and P. macerans DSM 24T was 34.0 ± 4.7 %. On the basis of phylogenetic, physiological and chemotaxonomic analysis data, strain WP-1T is considered to represent as a novel species of the genus Paenibacillus, for which the name Paenibacillus thermophilus sp. nov., is proposed, with the type strain WP-1T (=DSM 24746T = JCM 17693T = CCTCC AB 2011115T).



Similar content being viewed by others
References
Ash C, Priest FG, Collins MD (1993) Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test (Proposal for the creation of a new genus Paenibacillus). Antonie Van Leeuwenhoek 64:253–260
Behrendt U, Schumann P, Stieglmeier M, Pukall R, Augustin J, Spröer C, Schwendner P, Moissl-Eichinger C, Ulrich A (2010) Characterization of heterotrophic nitrifying bacteria with respiratory ammonification and denitrification activity-description of Paenibacillus uliginis sp. nov., an inhabitant of fen peat soil and Paenibacillus purispatii sp. nov., isolated from a spacecraft assembly clean room. Syst Appl Microbiol 33:328–336
Benardini JN, Vaishampayan PA, Schwendner P, Swanner E, Fukui Y, Osman S, Satomi M, Venkateswaran K (2011) Paenibacillus phoenicis sp. nov., isolated from the Phoenix Lander assembly facility and a subsurface molybdenum mine. Int J Syst Evol Microbiol 61:1338–1343
Cerny G (1978) Studies on aminopeptidase for the distinction of Gram-negative from Gram-positive bacteria. Eur J Appl Microbiol Biotechnol 5:113–122
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Cowan ST, Steel KJ (1965) Manual for the Identification of Medical Bacteria. Cambridge University Press, London
Cui XL, Mao PH, Zeng M, Li WJ, Zhang LP, Xu LH, Jiang CL (2001) Streptimonospora salina gen. nov., sp. nov., a new member of the family Nocardiopsaceae. Int J Syst Evol Microbiol 51:357–363
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Conference limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Heyndrickx M, Vandemeulebroecke K, Scheldeman P, Kersters K, De Vos P, Logan NA, Aziz AM, Ali N, Berkeley RCW (1996) A polyphasic reassessment of the genus Paenibacillus, reclassification of Bacillus lautus (Nakamura 1984) as Paenibacillus lautus comb. nov. and of Bacillus peoriae (Montefusco et al. 1993) as Paenibacillus peoriae comb. nov., and emended descriptions of P. lautus and of P. peoriae. Int J Syst Bacteriol 46:988–1003
Kämpfer P, Falsen E, Lodders N, Martin K, Kassmannhuber J, Busse H-J (2011) Paenibacillus chartarius sp. nov. isolated from a papermill. Int J Syst Evol Microbiol. doi:10.1099/ijs.0.035154-0
Khianngam S, Akaracharanya A, Tanasupawat S, Lee KC, Lee J-S (2009) Paenibacillus thailandensis sp. nov. and Paenibacillus nanensis sp. nov., xylanase-producing bacteria isolated from soil. Int J Syst Evol Microbiol 59:564–568
Komagata K, Suzuki K (1987) Lipids and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Kroppenstedt RM (1982) Separation of bacterial menaquinones by HPLC using reverse phase (RP 18) and a silver loaded ion exchanger as stationary phases. J Liquid Chromatogr 5:2359–2387
Lee M, Ten LN, Baek S-H, Im W-T, Aslam Z, Lee S-T (2007) Paenibacillus ginsengisoli sp. nov., a novel bacterium isolated from soil of a ginseng field in Pocheon province, South Korea. Antonie van Leeuwenhoek 91:127–135
Leifson E (1960) Atlas of Bacterial Flagellation. Academic Press, London
Logan NA, Clerck ED, Lebbe L, Verhelst A, Goris J, Forsyth G, Rodríguez-Díaz M, Heyndrickx M, De Vos P (2004) Paenibacillus cineris sp. nov. and Paenibacillus cookii sp. nov., from Antarctic volcanic soils and a gelatin-processing plant. Int J Syst Evol Microbiol 54:1071–1076
Logan NA, Berge O, Bishop AH, Busse H-J, De Vos P, Fritze D, Heyndrickx M, Kämpfer P, Rabinovitch L, Salkinoja-Salonen MS, Seldin L, Ventosa A (2009) Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int J Syst Evol Microbiol 59:2114–2121
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G + C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Osman S, Satomi M, Venkateswaran K (2006) Paenibacillus pasadenensis sp. nov. and Paenibacillus barengoltzii sp. nov., isolated from a spacecraft assembly facility. Int J Syst Evol Microbiol 56:1509–1514
Roux V, Raoult D (2004) Paenibacillus massiliensis sp. nov., Paenibacillus sanguinis sp. nov. and Paenibacillus timonensis sp. nov., isolated from blood cultures. Int J Syst Evol Microbiol 54:1049–1054
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Skerman VBD (1967) A Guide to the Identification of the Genera of Bacteria, 2nd edn. Williams & Wilkins, Baltimore
Smibert RM, Krieg NR (1981) General characterization. In: Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Philips GB (eds) Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC, pp 409–443
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tang SK, Tian XP, Zhi XY, Cai M, Wu JY, Yang LL, Xu LH, Li WJ (2008) Haloactinospora alba gen. nov., sp. nov., a halophilic filamentous actinomycete of the family Nocardiopsaceae. Int J Syst Evol Microbiol 58:2075–2080
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wang L, Baek S-H, Cui Y, Lee H-G, Lee S-T (2011) Paenibacillus sediminis sp. nov., a xylanolytic bacterium isolated from tidal flat adjacent to Ganghwa Island, Korea. Int J Syst Evol Microbiol. doi:10.1099/ijs.0.032102-0
Williams ST, Goodfellow M, Alderson G (1989) Genus Streptomyces Waksman and Henrici 1943, 339AL. In: Williams ST, Sharpe ME, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 4. Williams &Wilkins, Baltimore, pp. 2463–2468
Acknowledgments
The authors are grateful to Dr Hans-Peter Klenk of the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) for kindly providing reference type strain P. macerans DSM 24T. This work was supported by the National Natural Science Foundation of China (NSFC, 31000001). W-J Li was also supported by ‘Hundred Talents Program’ of the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, Y., Gao, S., Wei, DQ. et al. Paenibacillus thermophilus sp. nov., a novel bacterium isolated from a sediment of hot spring in Fujian province, China. Antonie van Leeuwenhoek 102, 601–609 (2012). https://doi.org/10.1007/s10482-012-9755-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-012-9755-6