Abstract
This is the first report of a catechol 1,2-dioxygenase from Stenotrophomonas maltophilia strain KB2 with high activity against catechol and its methyl derivatives. This enzyme was maximally active at pH 8.0 and 40 °C and the half-life of the enzyme at this temperature was 3 h. Kinetic studies showed that the value of K m and V max was 12.8 μM and 1,218.8 U/mg of protein, respectively. During our studies on kinetic properties of the catechol 1,2-dioxygenase we observed substrate inhibition at >80 μM. The nucleotide sequence of the gene encoding the S. maltophilia strain KB2 catechol 1,2-dioxygenase has high identity with other catA genes from members of the genus Pseudomonas. The deduced 314-residue sequence of the enzyme corresponds to a protein of molecular mass 34.5 kDa. This enzyme was inhibited by competitive inhibitors (phenol derivatives) only by ca. 30 %. High tolerance against condition changes is desirable in industrial processes. Our data suggest that this enzyme could be of use as a tool in production of cis,cis-muconic acid and its derivatives.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Adipic acid is an important industrial compound, production of which is necessary for the manufacture of nylon, polyurethane, insecticides and bactericides. One of the most important intermediates during synthesis of this acid is cis,cis-muconic acid, which easily converts to adipic acid by hydrogenation (Wu et al. 2006). Moreover, cis,cis-muconic acid has reactive carboxylate groups and a configuration of conjugated double bonds that can be useful as a raw material for new functional resins, pharmaceuticals and agrochemicals (Bang et al. 1996; Wingard and Finn 1969). However, there have been no secure and economical methods to synthesize cis,cis-muconic acid. Several reports have described the preparation of this compound starting with straight-chain compounds, however, the product has been the more thermodynamically stable trans,trans isomer and therefore in recent years biological methods have been used more often to produce cis,cis-muconic acid (Wingard and Finn 1969). For example, Schmidt and Knackmuss (1984) described Pseudomonas sp. strain B13, which produced cis,cis-muconate and 2-fluoro-cis,cis-muconate from benzoate and 3-fluorobenzoate. Kaneko et al. (2011) used recombinant Escherichia coli cells expressing the catA gene for high-yield production of cis,cis-muconic acid from catechol. This gene codes for catechol 1,2-dioxygenase (1,2-CTD), which plays a key role in catalysing aromatic ring cleavage at the ortho position to yield cis,cis-muconic acid.
Recently several catechol 1,2-dioxygenases, generally from members of the genera Arthrobacter, Acinetobacter, Pseudomonas, Sphingomonas and Rhodococcus have been described (Earhart et al. 2005; Guo et al. 2009; Guzik et al. 2011; Kim et al. 1998; Matera et al. 2010; Matsumura et al. 2004; Melo et al. 2010; Murakami et al. 1998; Nadaf and Ghosh 2011; Saxena and Thakur 2005; Wang et al. 2006). Most of them are enzymes with molecular mass 30–34 kDa, which consist of two either identical or non-identical subunits (Bruijnincx et al. 2008; Bugg 2003; Guzik et al. 2011; Patel et al. 1976; Vaillancourt et al. 2006). At the dimeric interface is located a hydrophobic cavity which is connected to phospholipid molecules (Matera et al. 2010). Nonheme iron in the ferric state is used as a cofactor for intradiol dioxygenases (Bruijnincx et al. 2008; Bugg 2003; Guzik et al. 2011; Patel et al. 1976; Vaillancourt et al. 2006). The iron is ligated by two histidines and two tyrosines. The initial coordination geometry is trigonal bipyramidal with tyrosine, histidine and a bound hydroxyl in the equatorial plane, and the other tyrosine and histidine as axial ligands (Earhart et al. 2005). The catalytic cycle of the intradiol dioxygenases involves binding of the catechol as a dianion, binding of dioxygen to the metal, in sequence formation of a peroxo and hydroperoxo intermediate. In the next step, the Criegee rearrangement occurs and O–O bond cleavage, which involves acyl migration to yield the cyclic anhydride and an iron-bound oxide or hydroxide, takes place. Hydrolysis of the anhydride leads to the formation of the final acyclic product (Bugg 2003; Bugg and Lin 2001; Vaillancourt et al. 2006; Vetting and Ohlendorf 2000).
Comprehensive studies on the substrate diversity and catalytic properties of catechol 1,2-dioxygenases are essential to facilitate the cheap and safe synthesis of cis,cis-muconic acid. Now we report novel a catechol 1,2-dioxygenase, characterized by high activity, isolated from Stenotrophomonas maltophilia strain KB2 which converted benzoic acid to cis,cis–muconic acid. We have identified the gene encoding the catechol 1,2-dioxygenase in S. maltophilia KB2 and deduced a putative three-dimensional structure of this enzyme from the amino acid sequence.
Materials and methods
Media and culture conditions
Stenotrophomonas maltophilia KB2 (VTT E-113197) was cultivated in mineral salts medium (MSM), as described previously (Wojcieszyńska et al. 2011) in the presence of 6 mM benzoic acid. Cultures were incubated at 30 °C and agitated at 130 rpm.
Preparation of cell extracts
Cells were harvested in the late exponential growth phase and centrifuged at 4,500×g for 15 min at 4 °C. Next, the cells were washed with 50 mM phosphate buffer, pH 7.0, and resuspended in the same buffer. Cells were sonicated 6× for 15 s and centrifuged at 9,000×g for 30 min at 4 °C. The supernatant was used as crude extract for enzyme assays.
Enzyme assays
Benzoic acid was used as the inducer of catechol 1,2-dioxygenase in the growth medium. Enzymatic activity of the enzyme was measured spectrophotometrically (Wojcieszyńska et al. 2011). After the addition of the enzyme, vials were incubated at 35 °C in a water-bath with shaking. At specific time intervals, 1 ml aliquots were withdrawn and used to monitor the reaction progress by measuring the product cis,cis-muconic acid at 260 nm. The extinction coefficient of the oxidation product of catechol was determined as ε260 nm = 16,800/M cm. One unit of enzyme activity was defined as the amount of enzyme required to generate 1 μmol of product per minute at 35 °C. The protein concentration was determined by the dye-binding procedure of Bradford using bovine serum albumin as a standard (Bradford 1976).
pH and temperature optima of catechol 1,2-dioxygenase
The effect of pH on the enzyme activity was determined by measuring the activity at 35 °C over the pH range 2.2–12.0 using the following buffers: 0.05 M glycine (pH 2.2), 0.05 M phosphate-citrate (pH 3.0–5.0), 0.05 M Sörensen (pH 6.0–8.0), 0.05 M borate (pH 9.0–10.0), and 0.05 Britton-Robinson (pH 11.00–12.00).
The optimum temperature was determined by assaying the enzyme activity at various temperatures (4–60 °C) in 50 mM phosphate buffer solution (pH 7.2).
Determination of kinetic constants of catechol 1,2-dioxygenase
The catalytic parameters (Michaelis–Menten constant, K m, and Maximum velocity, V max) for the enzyme were calculated by measuring the initial linear rates of the enzymatic reaction after the addition of different concentrations of catechol ranging from 0 to 100 μM at 35 °C. Three independent measurements were carried out for each substrate concentration. K m and V max were calculated from the Michaelis–Menten equation.
Substrate specificity
The impact of various substituted derivatives of aromatic compounds on enzyme activity was evaluated by incubating the enzyme with the respective aromatic compound (at 1 mM) for 3 min and assaying the activity. Dihydroxy-substituted derivatives of arene studied were 3- and 4-methylcatechol, 4,5- and 3,5-dichlorocatechol, and hydroquinone. The molar extinction coefficient used for the product from hydroquinone was 11,000/M cm (at 320 nm) (Kolvenbach et al. 2011; Spain and Gibson 1991). Catechol 1,2-dioxygenase activities for chlorinated and methylated derivatives of catechol were determined by the procedures of Dorn and Knackmuss (1978).
Activity in the presence of inhibitors
The impact of various phenols and chelators on enzyme activity was evaluated by incubating the enzyme with the respective inhibitor for 3 min and then assaying the residual activity. At specific time intervals, 1 ml aliquots were withdrawn and used to monitor the reaction progress by measuring the product cis,cis-muconic acid. Assay of catechol 1,2-dioxygenase was performed in the same way as in the case of non-inhibited enzyme. The phenols studied were 2-methylphenol, 3-methylphenol, 4-methylphenol, 2-chlorophenol, 4-chlorophenol, 2,4-dichlorophenol, each at 0.1, 0.2, and 0.3 mM concentration. Chelators studied were phenanthroline, EDTA, and 2,2′-dipirydyl (each at 1, 2, and 3 mM).
Amplification of catechol 1,2-dioxygenase gene
Genomic DNA, plasmid DNA isolation and other DNA manipulations were carried out according to established procedures (Sambrook et al. 1989). Oligonucleotides for the PCR were purchased from IBB PAN (Warsaw, Poland). Detection of the 1,2-CTD gene was carried out with primer designed by Guzik et al. 2011 i.e. 1,2D_zewF (GATGGATCCGTGAAAATTTCCCACATGC) and 1,2D_zewR (TGGATCCAGTAACGTTGCAGGTGCT). The PCR mixtures contained 0.5 μM of each primer, 10× Pfu buffer + MgSO4 (2 mM Mg2+), 10 mM K+, 3 % DMSO, 0.2 mM of each deoxynucleoside triphosphate, 1.25 U Pfu DNA polymerase (Sigma) and plasmid or chromosomal DNA as a template. For the 1,2-CTD genes, the annealing temperature was 61 °C (30 s) in the first 10 cycles followed by a step down to 59 °C (30 s) in the next 15 cycles, and 57 °C (30 s) in the last 15 cycles. Aliquots (10 μl) of the PCR products were analyzed by electrophoresis on a 1.0 % agarose gel stained with 0.5 ug/ml ethidium bromide. Gene sequencing was performed by using a Big DyeR Terminator Cycle Sequencing Kit (Applied Biosystem) and AbiPrism®3100 Genetic Analyzer. Computer analysis and processing of sequence information were performed by using Chromas LITE software (Technelysium Pty, Tewantin, Australia). The nucleotide sequence obtained for the catechol 1,2-dioxygenase gene from S. maltophilia strain KB2 has been deposited in the NCBI GenBank database under the accession number EU000397.1.
Molecular modeling of the catechol 1,2-dioxygenase enzyme
The amino acid sequence of the catechol 1,2-dioxygenase was deduced and followed by multiple sequence alignment using the CLC Free Workbench 6.3 software. The deduced structure of the catechol 1,2-dioxygenase was modeled using the interactive mode of the 3D-JIGSAW protein comparative modeling server (http://bmm.cancerresearchuk.org/~3djigsaw/). Structure models as x.pdb data files were analyzed using RasMol 2.6 software package.
Results and discussion
Production of cis,cis-muconic acid by catechol 1,2-dioxygenase
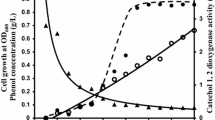
In last few decades enzymes with potential for usage in chemical synthesis at the industrial scale have been sought. It is especially important for production of stereoisomers since enzymes exhibit regioselectivity and stereoselectivity (Ran et al. 2008). Several studies have demonstrated production of cis,cis-muconic acid by microorganisms from benzene, toluene, benzoic acid or catechol (Bang and Choi 1995; Bang et al. 1996; Frost and Draths 1996, 1997). For example, a mutant strain of Arthrobacter sp. produced 44 g/l of cis,cis-muconic acid in two days of culture (Mizuno and Yoshikawa 1990). For the further enhancement of the cis,cis-muconic acid productivity, it is necessary to obtain high activity catechol 1,2-dioxygenase, the key enzyme in the cis,cis-muconate biosynthetic pathway (Kim et al. 1998; Wu et al. 2006). An earlier study showed that catechol 1,2-dioxygenase from S. maltophilia KB2 was observed after growing the strain in the presence of benzoate (Wojcieszyńska et al. 2011). We considered that the formation of this compound is dependent on substrate concentration. Figure 1 shows that the rate of cis,cis-muconic acid synthesis averaged 10 μM/min. The molar conversion yield based on the amount of consumed catechol was the theoretical value of 100 % (mol/mol). Similar results was obtained by Kaneko et al. (2011) during production of cis,cis-muconic acid by recombinant E. coli cells that expressed a catA gene from Pseudomonas putida mt-2.
Sequence analysis of the catechol 1,2-dioxygenase gene
Genes encoding catechol dioxygenases can be located on plasmids or/and on the chromosome (Neidle et al. 1998; Sauret-Ignazi et al. 1996;Vaillancourt et al. 2006; Wojcieszyńska et al. 2011). Our research indicated that strain KB2 contains plasmid DNA (Wojcieszyńska et al. 2011) and we thus performed PCR with chromosomal and plasmid DNA as a template. To amplify the catechol 1,2-dioxygenase S. maltophilia KB2 gene we used primers 1,2D_zewF and 1,2D_zewR (Guzik et al. 2011). A PCR product was obtained only with chromosomal DNA as a template, indicating that the gene is located on the chromosomal DNA. Sequencing of the PCR product resulted in a 1243 nucleotide sequence (deposited in GenBank under accession number EU000397). A phylogenetic tree was created (Fig. 2), based upon the catechol 1,2-dioxygenase gene sequences found in GenBank and the new sequence obtained in this study. There was high homology found with other intradiol dioxygenase genes.
Phylogenetic tree showing the position of the catechol 1,2-dioxygenase sequence from S. maltophilia KB2 in comparison with reference catechol 1,2-dioxygenases from other bacterial strains. The numbers at branch points indicate the confidence (in percent) as determined by bootstrap analysis with 100 replicates. The accession codes in the GenBank database and their origins are as follows: CP002290.1 for Pseudomonas putida BIRD-1 catechol 1,2-dioxygenase; CP002727.1 for Pseudomonas fulva 12-X catechol 1,2-dioxygenase; CP000094.2 for Pseudomonas fluorescens Pf0-1 catechol 1,2-dioxygenase; CP000926.1 for Pseudomonas putida GB-1 catechol 1,2-dioxygenase; CP002620.1 for Pseudomonas mendocina NK-01 catechol 1,2-dioxygenase; CT573326.1 for Pseudomonas entomophila L48 catechol 1,2-dioxygenase; AE015451.1 for Pseudomonas putida KT2440 catechol 1,2-dioxygenase; EU000397 for Stenotrophomonas maltophilia KB2 catechol 1,2-dioxygenase; D37782.1 for Pseudomonas putida_a catechol 1,2-dioxygenase; D37783.1 for Pseudomonas arvilla catechol 1,2-dioxygenase; AF363241.1 for Pseudomonas putida_b catechol 1,2-dioxygenase; CP000712.1 for Pseudomonas putida F1 catechol 1,2-dioxygenase
Structural properties of the catechol 1,2-dioxygenase
Knowledge of the catechol 1,2-dioxygenases 3-D structure can provide the important information into the molecular mechanisms of these enzymes. The deduced 314-residue amino acid sequence of S. maltophilia KB2 (deposited in the GenBank under accession number ABS86780.1) enzyme corresponds to a protein of molecular mass 34.5 kDa. Similar molecular weights for dioxygenase from Acinetobacter calcoaceticus and P. putida N6 were obtained by Neidle et al. (1998) and Guzik et al. (2011), respectively.
We predicted the 3-D structure of the catechol 1,2-dioxygenase from strain KB2 based on the deduced amino acid sequence by using the interactive mode of the 3D-JIGSAW protein comparative modeling server. Catechol 1,2-dioxygenase from strain KB2 (Fig. 3a) probably possesses an N-terminal domain with five α helices and a C-terminal domain consisting of β-sheets–structures typical for other intradiol dioxygenases as reported previously (Guzik et al. 2011; Ohlendorf et al. 1994; Vaillancourt et al. 1998). Similar molecular structure was also noted in another study of Pseudomonas arvilla C-1 catechol 1,2-dioxygenase, catechol 1,2-dioxygenase and 4-chlorocatechol 1,2-dioxygenase from Rhodococcus opacus 1CP (Earhart et al. 2005; Ferraroni et al. 2004; Matera et al. 2010). The α helices localized within N-terminal domain of the enzyme of strain C-1, like other known intradiol enzymes, were found to be involved in dimerization of enzyme subunits (Bugg 2003; Vaillancourt et al. 2006; Wojcieszyńska et al. 2011).
Intradiol dioxygenases coordinate ferric ion by two histidine, two tyrosine residues and one hydroxyl ion in a trigonalbipyramidal geometry (Bruijnincx et al. 2008; Bugg and Lin 2001; Earhart et al. 2005; Ferraroni et al. 2004; Matera et al. 2010; Melo et al. 2010). Within the active site of the R. opacus 1CP 4-chlorocatechol 1,2-dioxygenase, the coordination residues were identified at positions His-194, His-196, Tyr-134, and Tyr-169 (Ferraroni et al. 2004). The catechol 1,2-dioxygenase isolated from this same strain possess as iron ligands: Tyr-162, Tyr-196, His-220, and His-222 (Matera et al. 2010). Our work predicts His-226 Tyr-166, and Tyr-200 to be involved in ferric ions coordination (Figs. 3b, c, 4). However, comparison of the deduced amino acid sequence of the catechol 1,2-dioxygenase from S. maltophilia KB2 with other catechol 1,2-dioxygenases has shown that one of conserved active-site residues was altered in the strain KB2 sequence. We predict Gln-224 as a fourth ligand of iron ion (Fig. 4). Displacing one of the key iron bound ligands can cause changes in catalytic properties of enzyme and therefore we examined these in our study.
Multiple sequence alignment of predicted catechol 1,2-dioxygenase amino acid sequences carried out using CLC sequence viewer. Aligned sequences are 1,2-CTD of Acinetobacter calcoaceticus NCIB8250 (CAA85386.1), 1,2-CTD of Alcaligenes eutrophus CH34 (YP_587012.1), 1,2-CDT of Pseudomonas putida N6 (ABS86779.1), 1,2-CTD of Stenotrophomonas maltophilia KB2 (ABS86780.1), 1,2-CTD of Burkholderia sp. TH2 (BAC16779.1), CatA of Rhodococcus opacus 1CP (CAA67941.1), and 1,2-CTD of Rhodococcus erythropolis AN-13 (BAA11859.1). Residues in solid boxes indicate the Fe ligands
Kinetic properties of catechol 1,2-dioxygenase in S. maltophilia KB2 cell extracts
The pH-activity and temperature-activity curves showed that the maximum catechol 1,2-dioxygenase activity (3,062 U/mg protein) was at pH 8 and 40 °C, respectively (Fig. 5a, b). On the other hand, the examined enzyme was not very stable in this temperature (Fig. 5c). The half-life of the enzyme at 40 °C was 3 h (Fig. 5b). Interestingly, the enzyme lost 16.5 % of its enzymatic activity at 50 °C and the activity rapidly declined at 55 °C (Fig. 5b). A similar effect was observed by Wang et al. (2006) and Murakami et al. (1998) for catechol 1,2-dioxygenase from Pseudomonas aeruginosa and Arthrobacter species BA-5-17, respectively. The enzyme isolated from strain KB2 lost 100 % of its original enzymatic activity at pH 2.2 and about 83 % at pH 12.0 (Fig. 5a). The optimal pH of the catechol 1,2-dioxygenase is high compared with that of catechol 1,2-dioxygenase from Pseudomonas fluorescens, P. aeruginosa and Rhodococcus sp. NCIM2891 (Nadaf and Ghosh 2011; Saxena and Thakur 2005; Wang et al. 2006).
In order to calculate values of K m and V max parameters, the activity of the S. maltophilia KB2 catechol 1,2-dioxygenase was measured at different substrate concentrations as detailed in Materials and Methods. The K m and V max values obtained were 12.18 μM and 1,218.8 U/mg of protein, respectively (Fig. 5d). This V max value is notably higher (approximately 20–100-fold) than the activity of other catechol 1,2-dioxygenases. Catechol 1,2-dioxygenase form Acinetobacter radioresistens showed 25.8 U/mg of protein (Briganti et al. 2000). Suvorova et al. (2006) and Solyanikova et al. (2009) characterized catechol 1,2-dioxygenase from R. opacus 1CP and R. opacus 6a with activities of 9.6 U/mg of protein and 55.5 U/mg of protein, respectively. Of note, the K m value was 2-fold higher than those obtained by Wang et al. (2006) and Nadaf and Ghosh (2011). This result may therefore indicate lower affinity of enzyme to the substrate.
During our studies on kinetic properties of the catechol 1,2-dioxygenase, substrate inhibition at >80 μM was observed (Fig. 5d). In line with our results, Sauret-Ignazi et al. (1996) observed inhibition activity of an Alicaligenes eutrophus CH34 1,2-dioxygenase which catalyses tetrachlorocatechol degradation.
Influence of various substrates on catechol 1,2-dioxygenase activity in S. maltophilia KB2 cell extracts
Differences in substrate specificity is one of the interesting characteristics noted among the isofunctional dioxygenases from various sources. The relative activities of the catechol 1,2-dioxygenase from strain KB2 towards various substrates are given in Table 1. It was found that the enzyme showed activity against catechol, 3-methylcatechol, and 4-methylcatechol. No activity was observed for 3-chloro- and 4-chlorocatechol, 3,5-dichloro- and 4,5-dichlorocatechol and hydroquinone. It could be interpreted that a haloatom might prevent the dioxygenase from attacking the ring. Similar results were obtained by Briganti et al. (2000), Matsumura et al. (2004) and Murakami et al. (1998) for intradiol dioxygenases isolated from A. radioresistens, Rhodococcus sp. AN-22 and Arthrobacter species BA-5-17, respectively. Giedraityte and Kalediene (2009) reported only 27 and 6 % of the relative activity to catechol (7.42 U/mg of protein) of a catechol 1,2-dioxygenase from Geobacillus sp. towards 3-methylcatechol and 4-methylcatechol, respectively. Remarkably broader substrate specificity was described by Wang et al. (2006) and Guo et al. (2009) during characterization of catechol 1,2-dioxygenases from P. aeruginosa and Sphingomonas xenophaga QYY, respectively. The catechol 1,2-dioxygenase from R. opacus 1CP showed high activity against to catechol and methylcatechols. Moreover, this enzyme catalyzed cleavage of chlorocatechols, pyrogallol, hydroxyquinol, 2,3- and 3,4-dihydroxybenzoic acid ring (Matera et al. 2010).
Enzyme activity in S. maltophilia KB2 cell extracts in the presence of inhibitors
Phenols substituted in the ortho position, which structurally mimic catechols, are known as competitive inhibitors of catechol 1,2-dioxygenases because they coordinate to the iron (III) ion in the enzyme active site (Sauret-Ignazi et al. 1996; Vaillancourt et al. 1998). Most of the phenols studied affected enzyme activity at all tested concentrations (Table 2). Sauret-Ignazi et al. (1996) observed greater sensitivity of catechol 1,2-dioxygenase in the presence of para substituted phenols. However the catechol 1,2-dioxygenase from strain KB2 strain did not reveal dependence of activity changes on position and kind of substituents. A similar effect was observed by Kolomytseva et al. (2010) during analysis of the influence of monochloro- and monomethylphenols on activity of chlorocatechol 1,2-dioxygenases from Rhodococcus opacus 1CP.
The sensitivity of the catechol 1,2-dioxygenase from strain KB to both ferrous and ferric iron chelators (Table 2) may reflect the fact that the iron of the enzyme active site is more weakly bound than in the enzyme from Arthrobacter species BA-5-17 (Murakami et al. 1998), Trichosporon cutaneum (Varga and Neujahr 1970), P. aeruginosa (Wang et al. 2006) A. calcoaceticus (Patel et al. 1976), or Rhodococcus sp. AN-22 (Matsumura et al. 2004). Varga and Neujahr (1970) suggested a correlation between substrate specificity and the affinity of iron to catechol 1,2-dioxygenases. They reported that dioxygenases with strongly bound iron had narrow substrate specificity and vice versa. Our results are at variance with this suggestions since the catechol 1,2-dioxygenase from strain KB2 has a narrow specificity and apparently weakly bound iron. The sensitivity of our enzyme ton the chelators may be connected with the untypical ligand (Gln-224) of iron in the active site of the dioxygenase (Fig. 3c).
In conclusion catechol 1,2-dioxygenase from S. maltophilia strain KB2 could be a useful tool in the production of cis,cis-muconic acid and its derivatives due to its high activity. The high activity of the enzyme in the presence of methylcatechols may enables it to be used to produce methyl derivatives of cis,cis-muconic acid. Moreover, the temperature and pH tolerance, and resistance to competitive inhibitors, may be desirable features of the catechol 1,2-dioxygenase from KB2 strain for industrial processes .
References
Bang SG, Choi CY (1995) DO-stat fed-batch production of cis,cis-muconic acid from benzoic acid by Pseudomonas putida BM014. J Ferment Bioeng 79:381–383
Bang SG, Choi WJ, Choi CY, Cho MH (1996) Production of cis,cis-muconic acid from benzoic acid via microbial transformation. Biotechnol Bioprocess Eng 1:36–40
Bradford MM (1976) A rapid and sensitive method of the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–258
Briganti F, Pessione E, Giunta C, Mazzoli R, Scozzafava A (2000) Purification and catalytic properties of two catechol 1,2-dioxygenase isozymes from benzoate-grown cells of Acinetobacter radioresistens. J Protein Chem 19:709–716
Bruijnincx PCA, van Koten G, Gebbink RJMK (2008) Mononuclear non-heme iron enzymes with the 2-His-1-carboxylate facial triad: recent development in enzymology and modeling studies. Chem Soc Rev 37:2716–2744
Bugg TDH (2003) Dioxygenase enzymes: catalytic mechanisms and chemical models. Tetrahedron 59:7075–7101
Bugg TDH, Lin G (2001) Solving the riddle of the intradiol and extradiol catechol dioxygenases: how do enzymes control hydroperoxide rearrangements? Chem Commun 35:941–952
Dorn E, Knackmuss MJ (1978) Chemical structure and biodegradability of halogenated aromatic compounds—substituent effects on 1,2-dioxygenation of catechol. Biochem J 174:85–94
Earhart CA, Vetting MW, Gosu R, Michaud-Soret I, Que L, Ohlendorf DH (2005) Structure of catechol 1,2-dioxygenase from Pseudomonas arvilla. Biochem Biophys Res Commun 338:198–205
Ferraroni M, Solyanikova IP, Kolomytseva MP, Scozzafava A, Golovleva L, Briganti F (2004) Crystal structure of 4-chlorocatechol 1,2-dioxygenase from chlorophenol-utilizing Gram-positive Rhodococcus opacus 1CP. J Biol Chem 279:27646–27655
Frost JW, Draths KM (1996) Synthesis of adipic acid from biomass-derived carbon sources. United States Patent No. 5,487,987
Frost JW, Draths KM (1997) Bacterial cell transformants for production of cis,cis-muconic acid and catechol. United States Patent No. 5,616,496
Giedraityte G, Kalediene L (2009) Catechol 1,2-dioxygenase from α-naphthol degrading thermophilic Geobacillus sp. strain: purification and properties. Cent Eur J Biol 4:68–73
Guo M, Qu YY, Zhou JT, Li A, Uddin MS (2009) Characterization of catechol 1,2-dioxygenase from cell extracts of Sphingomonas xenophaga QYY. Sci China Ser B-Chem 52:615–620
Guzik U, Greń I, Hupert-Kocurek K, Wojcieszyńska D (2011) Catechol 1,2-dioxygenase from the new aromatic compounds—degrading Pseudomonas putida strain N6. Int Biodeter Biodegr 65:504–512
Kaneko A, Ishii Y, Kirimura K (2011) High-yield production of cis,cis-muconic acid from catechol in aqueous solution by biocatalyst. Chem Lett 40:381–383
Kim BJ, Choi WJ, Lee EY, Choi CY (1998) Enhancement of cis,cis-muconate productivity by overexpression of catechol 1,2-dioxygenase in Pseudomonas putida BCM114. Biotechnol Bioprocess Eng 3:112–114
Kolomytseva MP, Ferraroni M, Chernykh AM, Scozzafava A, Briganti F, Golovleva LA (2010) Experimental and theoretical affinity of substituted phenols to chlorocatechol 1,2-dioxygenases: a step toward the comprehension of inhibitor/substrate binding to intradiol dioxygenases. J Mol Catal B-Enzyme 64:53–59
Kolvenbach BA, Lenz M, Bendorf D, Rapp E, Fousek J, Vlcek C, Schaffer A, Gabriel FLP, Kohler H-PE, Corvini PFX (2011) Purification and characterization of hydroquinone dioxygenase from Sphingomonas sp. strain TTNP3. AMB Express 1:1–8
Matera I, Ferraroni M, Kolomytseva M, Golovleva L, Scozzafava A, Briganti F (2010) Catechol 1,2-dioxygenase from the gram-positive Rhodococcus opacus 1CP: quantitative structure/activity relationship and the crystal structures of native enzyme and catechol adducts. J Struct Biol 170:564–584
Matsumura E, Ooi S, Murakami S, Takenaka S, Aoki K (2004) Constitutive synthesis, purification, and characterization of catechol 1,2-dioxygenase from the aniline-assimilating bacterium Rhodococcus sp. AN-22. J Biosci Bioeng 98:71–76
Melo FA, Araujo APU, Costa-Filho AJ (2010) Role of cis,cis-muconic acid in the catalysis of Pseudomonas putida chlorocatechol 1,2-dioxygenase. Int J Biol Macromol 47:233–237
Mizuno S, Yoshikawa N (1990) Microbial production of cis,cis-muconic acid. J Biotechnol 14:203–210
Murakami S, Wang CL, Naito A, Shinke R, Aoki K (1998) Purification and characterization of four catechol 1,2-dioxygenase isozymes from the benzamide-assimilating bacterium Arthrobacter species BA-5-17. Microbiol Res 153:163–171
Nadaf NH, Ghosh JS (2011) Purification and characterization of catechol 1,2-dioxygenase from Rhodocococcus sp. NCIM 2891. Res J Environ Earth Sci 3:608–613
Neidle EL, Hartnett Ch, Bonitz S, Ornston LN (1998) DNA sequence of the Acinetobacter calcoaceticus catechol 1,2-dioxygenase I structural gene catA: evidence for evolutionary divergence of intradiol dioxygenases by acquisition of DNA sequence repetitions. J Bacteriol 170:4874–4880
Ohlendorf DH, Orville AM, Lipscomb JD (1994) Structure of protocatechuate 3,4-dioxygenase from Pseudomonas aeruginosa at 2.15 Å resolution. J Mol Biol 244:586–608
Patel RN, Hou CT, Felix A, Lillard MO (1976) Catechol 1,2-dioxygenase from Acinetobacter calcoaceticus: purification and properties. J Bacteriol 127:536–544
Ran N, Zhao L, Chen Z, Tao J (2008) Recent applications of biocatalysis in developing green chemistry for chemical synthesis at the industrial scale. Green Chem 10:361–372
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Sauret-Ignazi G, Gagnon J, Béguin J, Barrelle M, Markowicz Y, Pelmont J, Toussaint A (1996) Characterisation of chromosomally encoded catechol 1,2-dioxygenase (E.C. 1.13.11.1) from Alcaligenes eutrophus CH34. Arch Microbiol 166:42–50
Saxena P, Thakur S (2005) Purification and characterization of catechol 1,2-dioxygenase of Pseudomonas fluorescens for degradation of chlorobenzoic acid. Indian J Biotechnol 4:134–138
Schmidt E, Knackmuss H-J (1984) Production of cis,cis-muconate from benzoate and fluoro-cis,cis-muconate from 3-fluorobenzoate by benzoate degrading bacteria. Appl Microbiol Biotechnol 20:351–355
Solyanikova IP, Konovalova EI, Golovleva LA (2009) Methylcatechol 1,2-dioxygenase of Rhodococcus opacus 6a is a new type of the catechol-cleaving enzyme. Biochemistry (Moscow) 74:994–1001
Spain JC, Gibson DT (1991) Pathway for biodegradation of p-nitrophenol in a Moraxella sp. Appl Environ Microbiol 57:812–819
Suvorova MM, Solyanikova IP, Golovleva LA (2006) Specificity of catechol ortho-cleavage during para-toluate degradation by Rhodococcus opacus 1CP. Biochemistry (Moscow) 71:1316–1323
Vaillancourt FH, Han S, Fortin PD, Bolin JT, Eltis LD (1998) Molecular basis for the stabilization and inhibition of 2,3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol. J Biol Chem 273:34887–34895
Vaillancourt FH, Bolin JT, Eltis LD (2006) The ins and outs of ring-cleaving dioxygenases. Crit Rev Biochem Mol Biol 41:241–267
Varga JM, Neujahr HY (1970) Purification and properties of catechol 1,2-oxygenase from Trichosporon cutaneum. Eur J Biochem 12:427–434
Vetting MW, Ohlendorf DH (2000) The 1.8 Å crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker. Structure 8:429–440
Wang C-L, You S-L, Wang S-L (2006) Purification and characterization of a novel catechol 1,2-dioxygenase from Pseudomonas aeruginosa with benzoic acid as a carbon source. Process Biochem 41:1594–1601
Wingard LB, Finn RK (1969) Oxidation of catechol to cis,cis-muconic acid with ozone. I&EC Product Res Dev 8:65–70
Wojcieszyńska D, Guzik U, Greń I, Perkosz M, Hupert-Kocurek K (2011) Induction of aromatic ring–cleavage dioxygenases in Stenotrophomonas maltophilia strain KB2 in cometabolic systems. World J Microbiol Biotechnol 27:805–811
Wu C-M, Wu C-CSuC-C, Lee S-N, Lee Y-A, Wu J-Y (2006) Micriobial synthesis of cis,cis-muconic acid from benzoate by Sphingobacterium sp. mutants. Biochem Eng J 29:35–40
Acknowledgments
We thank Dr Renata Zub (Department of Molecular Biology, Institute of Oncology, Warsaw, Poland) for DNA sequencing. Aleksandra Bury is acknowledged for her excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Guzik, U., Hupert-Kocurek, K., Sitnik, M. et al. High activity catechol 1,2-dioxygenase from Stenotrophomonas maltophilia strain KB2 as a useful tool in cis,cis-muconic acid production. Antonie van Leeuwenhoek 103, 1297–1307 (2013). https://doi.org/10.1007/s10482-013-9910-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-013-9910-8