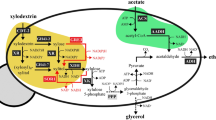
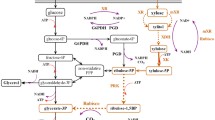
Abstract
Accumulation of reduced byproducts such as glycerol and xylitol during xylose fermentation by engineered Saccharomyces cerevisiae hampers the economic production of biofuels and chemicals from cellulosic hydrolysates. In particular, engineered S. cerevisiae expressing NADPH-linked xylose reductase (XR) and NAD+-linked xylitol dehydrogenase (XDH) produces substantial amounts of the reduced byproducts under anaerobic conditions due to the cofactor difference of XR and XDH. While the additional expression of a water-forming NADH oxidase (NoxE) from Lactococcus lactis in engineered S. cerevisiae with the XR/XDH pathway led to reduced glycerol and xylitol production and increased ethanol yields from xylose, volumetric ethanol productivities by the engineered yeast decreased because of growth defects from the overexpression of noxE. In this study, we introduced noxE into an engineered yeast strain (SR8) exhibiting near-optimal xylose fermentation capacity. To overcome the growth defect caused by the overexpression of noxE, we used a high cell density inoculum for xylose fermentation by the SR8 expressing noxE. The resulting strain, SR8N, not only showed a higher ethanol yield and lower byproduct yields, but also exhibited a high ethanol productivity during xylose fermentation. As noxE overexpression elicits a negligible growth defect on glucose conditions, the beneficial effects of noxE overexpression were substantial when a mixture of glucose and xylose was used. Consumption of glucose led to rapid cell growth and therefore enhanced the subsequent xylose fermentation. As a result, the SR8N strain produced more ethanol and fewer byproducts from a mixture of glucose and xylose than the parental SR8 strain without noxE overexpression. Our results suggest that the growth defects from noxE overexpression can be overcome in the case of fermenting lignocellulose-derived sugars such as glucose and xylose.





Similar content being viewed by others
References
Ansell R, Granath K, Hohmann S, Thevelein JM, Adler L (1997) The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J 16:2179–2187. doi:10.1093/emboj/16.9.2179
Bengtsson O, Hahn-Hagerdal B, Gorwa-Grauslund MF (2009) Xylose reductase from Pichia stipitis with altered coenzyme preference improves ethanolic xylose fermentation by recombinant Saccharomyces cerevisiae. Biotechnol Biofuels 2:9. doi:10.1186/1754-6834-2-9 (pii: 1754-6834-2-9)
Brat D, Boles E, Wiedemann B (2009) Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl Environ Microbiol 75:2304–2311. doi:10.1128/AEM.02522-08 (pii: AEM.02522-08)
Bro C, Regenberg B, Forster J, Nielsen J (2006) In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab Eng 8:102–111. doi:10.1016/j.ymben.2005.09.007 (pii: S1096-7176(05)00078-9)
Eliasson A, Christensson C, Wahlbom CF, Hahn-Hagerdal B (2000) Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol 66:3381–3386
Grotkjaer T, Christakopoulos P, Nielsen J, Olsson L (2005) Comparative metabolic network analysis of two xylose fermenting recombinant Saccharomyces cerevisiae strains. Metab Eng 7:437–444. doi:10.1016/j.ymben.2005.07.003 (pii: S1096-7176(05)00057-1)
Harhangi HR, Akhmanova AS, Emmens R, van der Drift C, de Laat WT, van Dijken JP, Jetten MS, Pronk JT, Op den Camp HJ (2003) Xylose metabolism in the anaerobic fungus Piromyces sp. strain E2 follows the bacterial pathway. Arch Microbiol 180:134–141. doi:10.1007/s00203-003-0565-0
Heux S, Cachon R, Dequin S (2006) Cofactor engineering in Saccharomyces cerevisiae: expression of a H2O-forming NADH oxidase and impact on redox metabolism. Metab Eng 8:303–314. doi:10.1016/j.ymben.2005.12.003 (pii: S1096-7176(05)00114-X)
Ho NW, Chen Z, Brainard AP (1998) Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Environ Microbiol 64:1852–1859
Hoefnagel MH, Starrenburg MJ, Martens DE, Hugenholtz J, Kleerebezem M, Van S II, Bongers R, Westerhoff HV, Snoep JL (2002) Metabolic engineering of lactic acid bacteria, the combined approach: kinetic modelling, metabolic control and experimental analysis. Microbiology 148:1003–1013
Hou J, Shen Y, Li XP, Bao XM (2007) Effect of the reversal of coenzyme specificity by expression of mutated Pichia stipitis xylitol dehydrogenase in recombinant Saccharomyces cerevisiae. Lett Appl Microbiol 45:184–189. doi:10.1111/j.1472-765X.2007.02165.x (pii: LAM2165)
Hou J, Suo F, Wang C, Li X, Shen Y, Bao X (2014) Fine-tuning of NADH oxidase decreases byproduct accumulation in respiration deficient xylose metabolic Saccharomyces cerevisiae. BMC Biotechnol 14:13. doi:10.1186/1472-6750-14-13
Hou J, Vemuri GN, Bao X, Olsson L (2009) Impact of overexpressing NADH kinase on glucose and xylose metabolism in recombinant xylose-utilizing Saccharomyces cerevisiae. Appl Microbiol Biotechnol 82:909–919. doi:10.1007/s00253-009-1900-4
Jeffries TW (2006) Engineering yeasts for xylose metabolism. Curr Opin Biotechnol 17:320–326. doi:10.1016/j.copbio.2006.05.008 (pii: S0958-1669(06)00066-8)
Jeppsson M, Bengtsson O, Franke K, Lee H, Hahn-Hagerdal B, Gorwa-Grauslund MF (2006) The expression of a Pichia stipitis xylose reductase mutant with higher K(M) for NADPH increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Biotechnol Bioeng 93:665–673. doi:10.1002/bit.20737
Jin YS, Jeffries TW (2003) Changing flux of xylose metabolites by altering expression of xylose reductase and xylitol dehydrogenase in recombinant Saccharomyces cerevisiae. Appl Biochem Biotechnol 105–108:277–286 (pii: ABAB-106-1-3-277)
Johansson B, Hahn-Hagerdal B (2002) Overproduction of pentose phosphate pathway enzymes using a new CRE-loxP expression vector for repeated genomic integration in Saccharomyces cerevisiae. Yeast 19:225–231. doi:10.1002/yea.833
Kim SR, Ha S-J, Wei N, Oh EJ, Jin Y-S (2012) Simultaneous co-fermentation of mixed sugars: a promising strategy for producing cellulosic ethanol. Trends Biotechnol 30:274–282. doi:10.1016/j.tibtech.2012.01.005
Kim SR, Skerker JM, Kang W, Lesmana A, Wei N, Arkin AP, Jin YS (2013) Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae. PLoS One 8:e57048. doi:10.1371/journal.pone.0057048
Kurtzman CP, Suzuki M (2010) Phylogenetic analysis of ascomycete yeasts that form coenzyme Q-9 and the proposal of the new genera Babjeviella, Meyerozyma, Millerozyma, Priceomyces, and Scheffersomyces. Mycoscience 51:2–14. doi:10.1007/s10267-009-0011-5
Kuyper M, Harhangi HR, Stave AK, Winkler AA, Jetten MS, de Laat WT, den Ridder JJ, Op den Camp HJ, van Dijken JP, Pronk JT (2003) High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res 4:69–78 (pii: S1567135603001417)
Kuyper M, Hartog MM, Toirkens MJ, Almering MJ, Winkler AA, van Dijken JP, Pronk JT (2005) Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res 5:399–409. doi:10.1016/j.femsyr.2004.09.010
Lau MW, Gunawan C, Balan V, Dale BE (2010) Comparing the fermentation performance of Escherichia coli KO11, Saccharomyces cerevisiae 424A(LNH-ST) and Zymomonas mobilis AX101 for cellulosic ethanol production. Biotechnol Biofuels 3:11. doi:10.1186/1754-6834-3-11
Lee SH, Kodaki T, Park YC, Seo JH (2012) Effects of NADH-preferring xylose reductase expression on ethanol production from xylose in xylose-metabolizing recombinant Saccharomyces cerevisiae. J Biotechnol 158:184–191. doi:10.1016/j.jbiotec.2011.06.005
Liang L, Zhang J, Lin Z (2007) Altering coenzyme specificity of Pichia stipitis xylose reductase by the semi-rational approach casting. Microb Cell Fact 6:36. doi:10.1186/1475-2859-6-36 (pii: 1475-2859-6-36)
Liu EK, Hu Y (2010) Construction of a xylose-fermenting Saccharomyces cerevisiae strain by combined approaches of genetic engineering, chemical mutagenesis and evolutionary adaptation. Biochem Eng J 48:204–210. doi:10.1016/j.bej.2009.10.011
Lopez de Felipe F, Kleerebezem M, de Vos WM, Hugenholtz J (1998) Cofactor engineering: a novel approach to metabolic engineering in Lactococcus lactis by controlled expression of NADH oxidase. J Bacteriol 180:3804–3808
Lu C, Jeffries T (2007) Shuffling of promoters for multiple genes to optimize xylose fermentation in an engineered Saccharomyces cerevisiae strain. Appl Environ Microbiol 73:6072–6077. doi:10.1128/AEM.00955-07
Matsushika A, Inoue H, Kodaki T, Sawayama S (2009) Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl Microbiol Biotechnol 84:37–53. doi:10.1007/s00253-009-2101-x
Matsushika A, Inoue H, Watanabe S, Kodaki T, Makino K, Sawayama S (2009) Efficient bioethanol production by a recombinant flocculent Saccharomyces cerevisiae strain with a genome-integrated NADP+-dependent xylitol dehydrogenase gene. Appl Environ Microbiol 75:3818–3822. doi:10.1128/AEM.02636-08 (pii: AEM.02636-08)
Matsushika A, Watanabe S, Kodaki T, Makino K, Inoue H, Murakami K, Takimura O, Sawayama S (2008) Expression of protein engineered NADP+-dependent xylitol dehydrogenase increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Appl Microbiol Biotechnol 81:243–255. doi:10.1007/s00253-008-1649-1
Olsson L, Hahn-Hägerdal B (1996) Fermentation of lignocellulosic hydrolysates for ethanol production. Enzym Microb Tech 18:312–331. doi:10.1016/0141-0229(95)00157-3
Parekh RN, Shaw MR, Wittrup KD (1996) An integrating vector for tunable, high copy, stable integration into the dispersed Ty delta sites of Saccharomyces cerevisiae. Biotechnol Prog 12:16–21. doi:10.1021/bp9500627
Petschacher B, Nidetzky B (2008) Altering the coenzyme preference of xylose reductase to favor utilization of NADH enhances ethanol yield from xylose in a metabolically engineered strain of Saccharomyces cerevisiae. Microb Cell Fact 7:9. doi:10.1186/1475-2859-7-9 (pii: 1475-2859-7-9)
Rizzi M, Harwart K, Erlemann P, Bui-Thanh N-A, Dellweg H (1989) Purification and properties of the NAD+-xylitol-dehydrogenase from the yeast Pichia stipitis. J Ferment Bioeng 67:20–24. doi:10.1016/0922-338X(89)90080-9
Runquist D, Hahn-Hagerdal B, Bettiga M (2010) Increased ethanol productivity in xylose-utilizing Saccharomyces cerevisiae via a randomly mutagenized xylose reductase. Appl Environ Microbiol 76:7796–7802. doi:10.1128/AEM.01505-10 (pii: AEM.01505-10)
Schiestl RH, Gietz RD (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16:339–346
Sonderegger M, Jeppsson M, Larsson C, Gorwa-Grauslund MF, Boles E, Olsson L, Spencer-Martins I, Hahn-Hagerdal B, Sauer U (2004) Fermentation performance of engineered and evolved xylose-fermenting Saccharomyces cerevisiae strains. Biotechnol Bioeng 87:90–98. doi:10.1002/bit.20094
Sun Y, Cheng J (2002) Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol 83:1–11. doi:10.1016/s0960-8524(01)00212-7
Toivari MH, Aristidou A, Ruohonen L, Penttila M (2001) Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae: importance of xylulokinase (XKS1) and oxygen availability. Metab Eng 3:236–249. doi:10.1006/mben.2000.0191S1096-7176(00)90191-5
Vemuri GN, Eiteman MA, McEwen JE, Olsson L, Nielsen J (2007) Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 104:2402–2407. doi:10.1073/pnas.0607469104 (pii: 0607469104)
Verduyn C, Van Kleef R, Frank J, Schreuder H, Van Dijken JP, Scheffers WA (1985) Properties of the NAD(P)H-dependent xylose reductase from the xylose-fermenting yeast Pichia stipitis. Biochem J 226:669–677
Verho R, Londesborough J, Penttila M, Richard P (2003) Engineering redox cofactor regeneration for improved pentose fermentation in Saccharomyces cerevisiae. Appl Environ Microbiol 69:5892–5897
Walfridsson M, Bao X, Anderlund M, Lilius G, Bulow L, Hahn-Hagerdal B (1996) Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl Environ Microbiol 62:4648–4651
Watanabe S, Abu Saleh A, Pack SP, Annaluru N, Kodaki T, Makino K (2007) Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology 153:3044–3054. doi:10.1099/mic.0.2007/007856-0 (pii: 153/9/3044)
Watanabe S, Kodaki T, Makino K (2005) Complete reversal of coenzyme specificity of xylitol dehydrogenase and increase of thermostability by the introduction of structural zinc. J Biol Chem 280:10340–10349. doi:10.1074/jbc.M409443200 (pii: M409443200)
Watanabe S, Pack SP, Saleh AA, Annaluru N, Kodaki T, Makino K (2007) The positive effect of the decreased NADPH-preferring activity of xylose reductase from Pichia stipitis on ethanol production using xylose-fermenting recombinant Saccharomyces cerevisiae. Biosci Biotechnol Biochem 71:1365–1369 (pii: JST.JSTAGE/bbb/70104)
Wisselink HW, Toirkens MJ, Wu Q, Pronk JT, van Maris AJ (2009) Novel evolutionary engineering approach for accelerated utilization of glucose, xylose, and arabinose mixtures by engineered Saccharomyces cerevisiae strains. Appl Environ Microbiol 75:907–914. doi:10.1128/AEM.02268-08 (pii: AEM.02268-08)
Wyman CE (2007) What is (and is not) vital to advancing cellulosic ethanol. Trends Biotechnol 25:153–157. doi:10.1016/j.tibtech.2007.02.009
Zhang GC, Kong II, Wei N, Peng D, Turner TL, Sung BH, Sohn JH, Jin YS (2016) Optimization of an acetate reduction pathway for producing cellulosic ethanol by engineered yeast. Biotechnol Bioeng. doi:10.1002/bit.26021
Zhang GC, Liu JJ, Ding WT (2012) Decreased xylitol formation during xylose fermentation in Saccharomyces cerevisiae due to overexpression of water-forming NADH oxidase. Appl Environ Microbiol 78:1081–1086. doi:10.1128/AEM.06635-11 (pii: AEM.06635-11)
Zhang GC, Liu JJ, Kong II, Kwak S, Jin YS (2015) Combining C6 and C5 sugar metabolism for enhancing microbial bioconversion. Curr Opin Chem Biol 29:49–57. doi:10.1016/j.cbpa.2015.09.008
Acknowledgements
This work was supported by funding from the Energy Biosciences Institute and TLT would like to acknowledge that this project was supported by the Agriculture and Food Research Initiative Competitive Grant No. 2015-67011-22806 from the USDA National Institute of Food and Agriculture.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, GC., Turner, T.L. & Jin, YS. Enhanced xylose fermentation by engineered yeast expressing NADH oxidase through high cell density inoculums. J Ind Microbiol Biotechnol 44, 387–395 (2017). https://doi.org/10.1007/s10295-016-1899-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1899-3